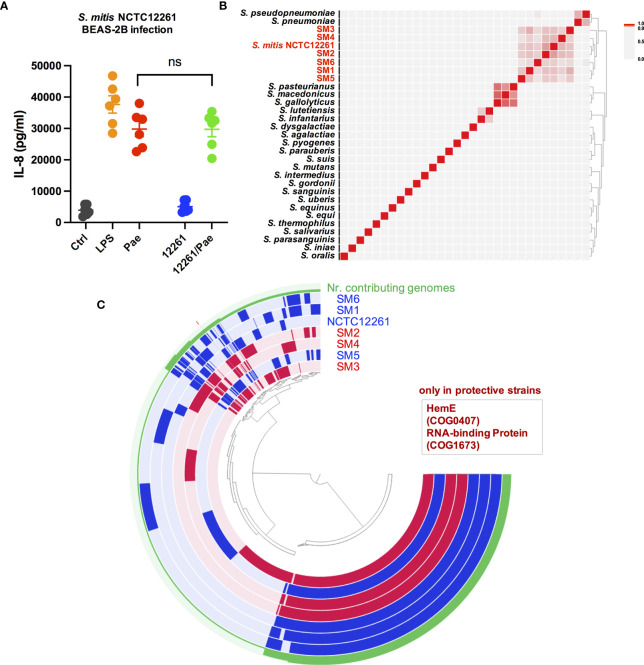
Figure 4.
Pan-genome analysis reveals genes with specific functions only present in the protective strains SM2, SM3 and SM4. (A) Co-infection of S. mitis type strain NCTC12261(labelled as 12261) and P. aeruginosa PAO1 (Pae) in BEAS-2B cells could not reduce the IL-8 production triggered by P. aeruginosa infection. ns, not significant. (B) ANIm analysis of complete whole genome sequences of Streptococcus type strains with SM1, SM2, SM3, SM4, SM5 and SM6. A heatmap and dendrogram of ANIm between these genomes is shown. (C) Pan-genome analysis to compare strains with protective effects and strains without protective effects. One layer represents one genome, with one extra layer showing number of contributing genomes to each orthologous gene family (Nr. contributing genomes). Genomes of strains showing protective effects (SM2, SM3, SM4) are colored in red, and genomes of strains without protective effects (NCTC12261, SM1, SM5 and SM6) are colored in blue. The clustering is based on presence/absence of orthologous gene family using Euclidean distance and Ward Linkage. Two genes with function annotation COG0407 and COG1673 are only detected in strains with protective effects, while missing in strains without protective effects.