Figure 3.
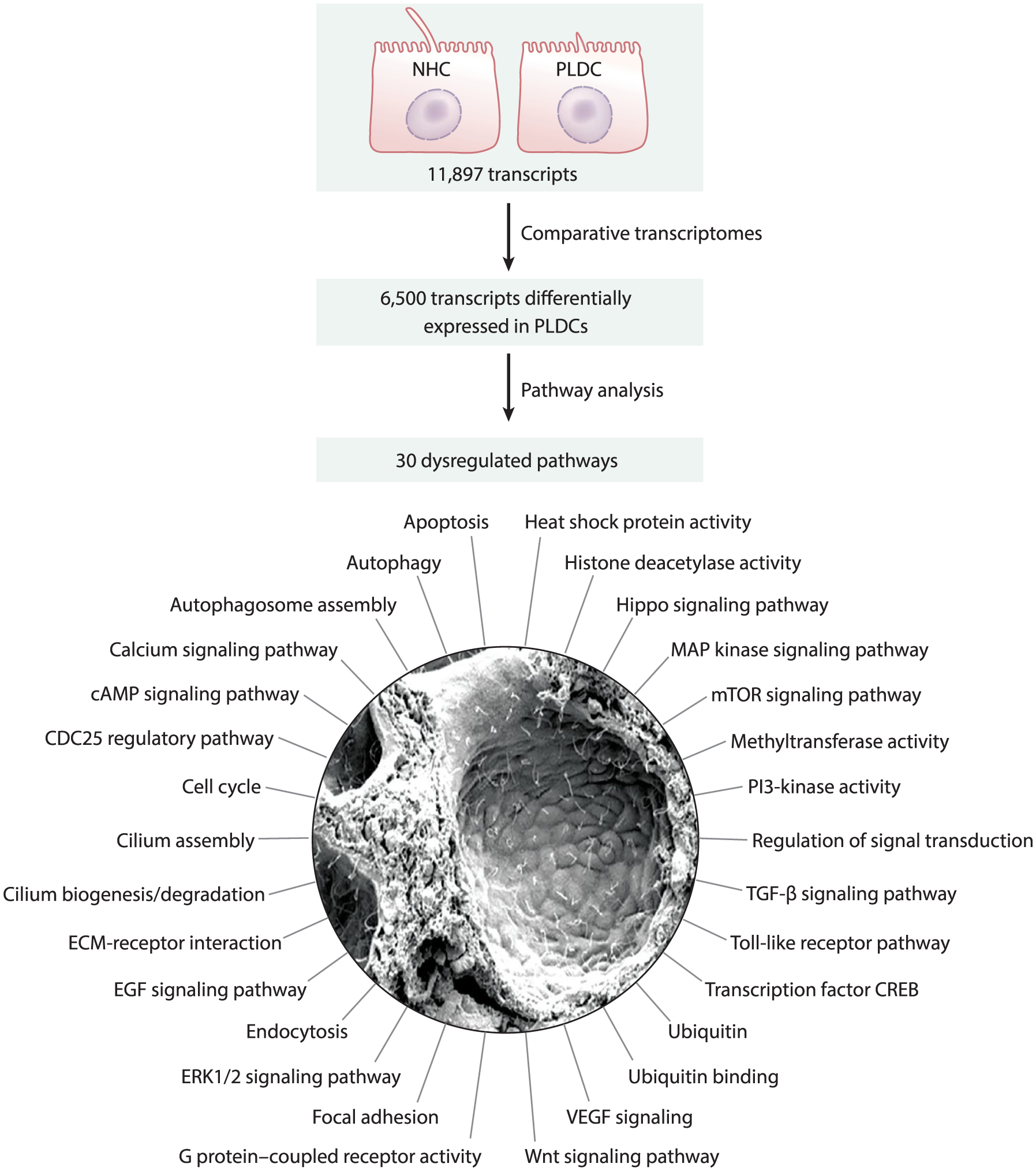
Pathways dysregulated in PLD. Transcriptome profiling of NHCs and PLDCs revealed that out of 11,897 transcripts present in both cell lines, 6,500 transcripts were differentially expressed in PLDCs (i.e., up- or downregulated). Functional annotation clustering of these transcripts by the Kyoto Encyclopedia of Genes and Genomes pathways indicates 30 altered pathways in PLDCs. Abbreviations: cAMP, cyclic adenosine monophosphate; CDC25, cell division cycle 25; CREB, cAMP-response element-binding protein; ECM, extracellular matrix; EGF, endothelial growth factor; ERK, extracellular signal-regulated kinase; MAP, mitogen-activated protein; mTOR mammalian target of rapamycin; NHC, normal human cholangiocyte; PI3, phosphoinositide 3; PLD, polycystic liver disease; PLDC, polycystic liver disease cholangiocyte; TGF, transforming growth factor; VEGF, vascular endothelial growth factor. Scanning electron microscope image adapted with permission from Reference 22.
