Fig. 1: Overview of the VascuViz pipeline for multimodality 3D vascular imaging and multiscale data integration:
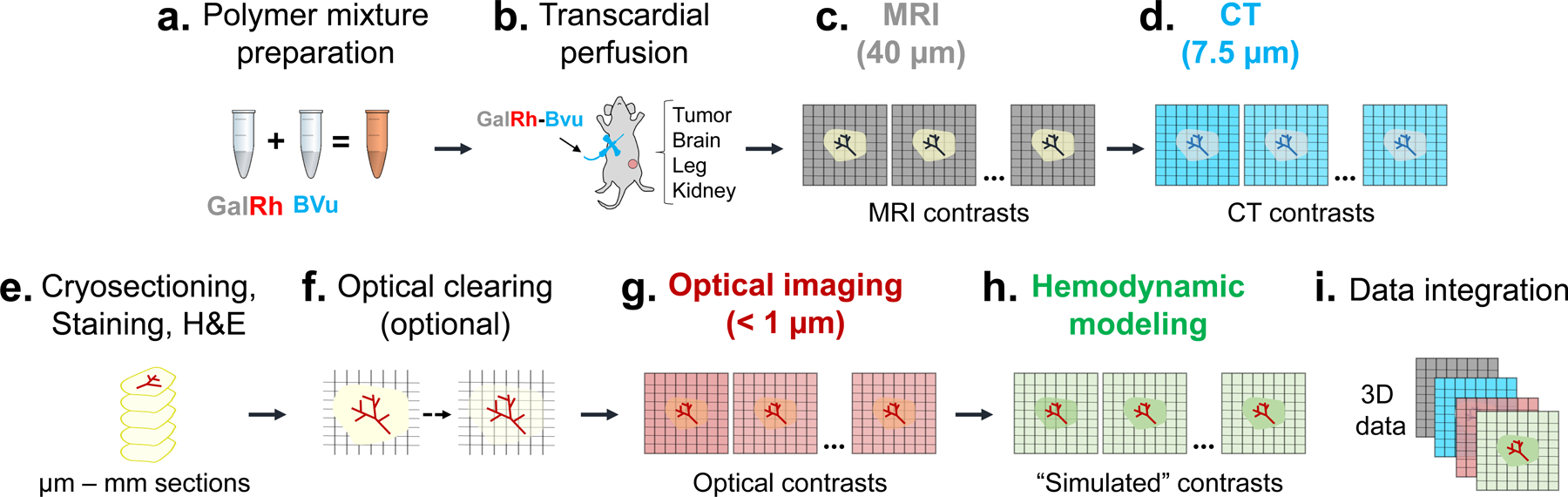
(a) First, the GalRh-BVu mixture was prepared by combining a fluorescently conjugated MRI contrast agent Galbumin™-Rhodamine (GalRh) with a radio-opqaue BriteVu® (BVu) solution (~30 min). (b) Next, vascular labeling was achieved via transcardial perfusion of the GalRh-BVu mixture (~15 min). After perfusion fixation, tumor and healthy tissues (e.g. brain, kidney and hind limb) were excised and immersion fixed overnight. (c) Then, T1-weighted (T1W) and diffusion-weighted (DW) MRI images were acquired at 40 μm and 100 μm isotropic (i.e. macroscopic) spatial resolution, respectively. (d) CT contrasts were acquired from the same sample at 7.5–9 μm isotropic (i.e. mesoscopic) spatial resolution. (e) Then, the sample was sectioned into 10 μm to 1.5 mm slices and immunohistochemical (IHC) or Hematoxylin and Eosin (H&E) staining performed. (f) If required, thicker tissue sections (i.e. > 1 mm) could be optically cleared at this stage. (g) Optical imaging was performed using either 3D light-sheet microscopy (LSM), multiphoton microscopy (MPM) or second harmonic generation (SHG) imaging at 0.5–5 μm lateral and 1–3 μm axial (i.e. microscopic) spatial resolution. (h) Following this step, hemodyamic modeling was performed using high-resolution 3D microvascular network data and additional “simulated” contrasts (e.g. blood flow) generated. (i) Finally, multicontrast data from (c-g) were processed and integrated into 3D data volumes for different vascular systems biology applications.
