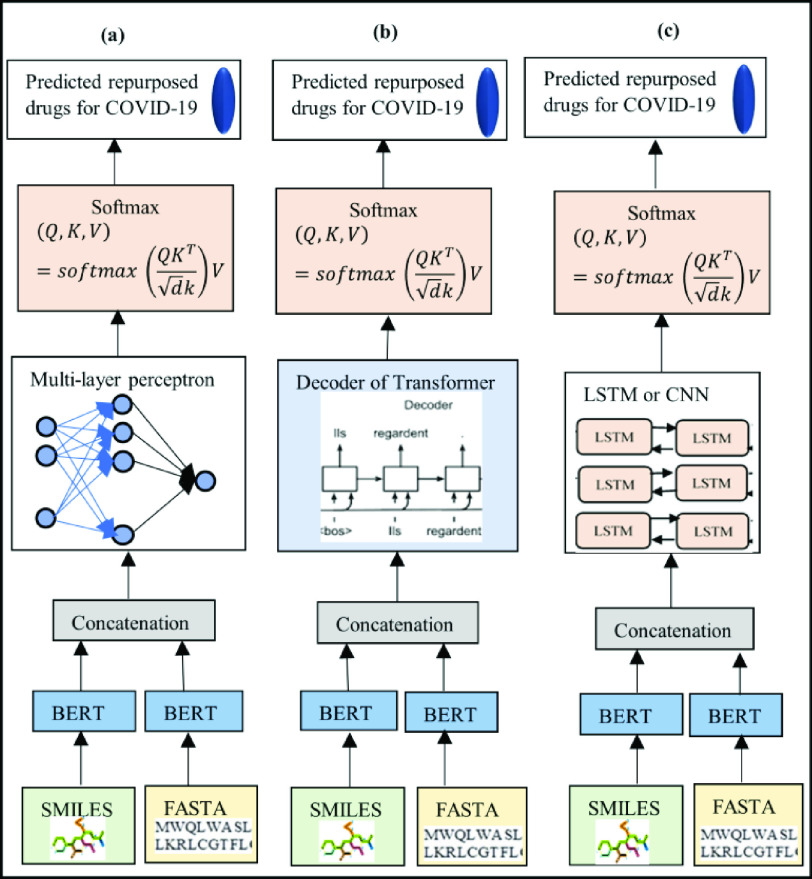
Fig. 6.
Illustration of four proposed encoder–decoder frameworks for predicting repurposed drug candidates for COVID-19. The two inputs are represented by molecular structures and protein sequences. After training the task-specific datasets using pretrained BERT-based models, the outputs from the pretrained encoders are concatenated and directed to a decoder. (a) Two inputs, drug molecules (SMILES) and protein molecules (FASTA), which are processed by BERT. The outputs are then concatenated before processed using multilayer perceptron with an activated softmax to predict repurposed drug candidates for COVID-19. (b) Two inputs, drug molecules (SMILES) and protein molecules (FASTA). The two inputs are processed by BERT and the outputs are concatenated before processed by transformer with activated softmax to predict repurposed drugs. (c) Two inputs, drug molecules (SMILES) and protein molecules (FASTA). The two inputs are processed by BERT and the outputs are then concatenated before processed using either LSTM or CNN with activated softmax to predict repurposed drug candidates for COVID-19.