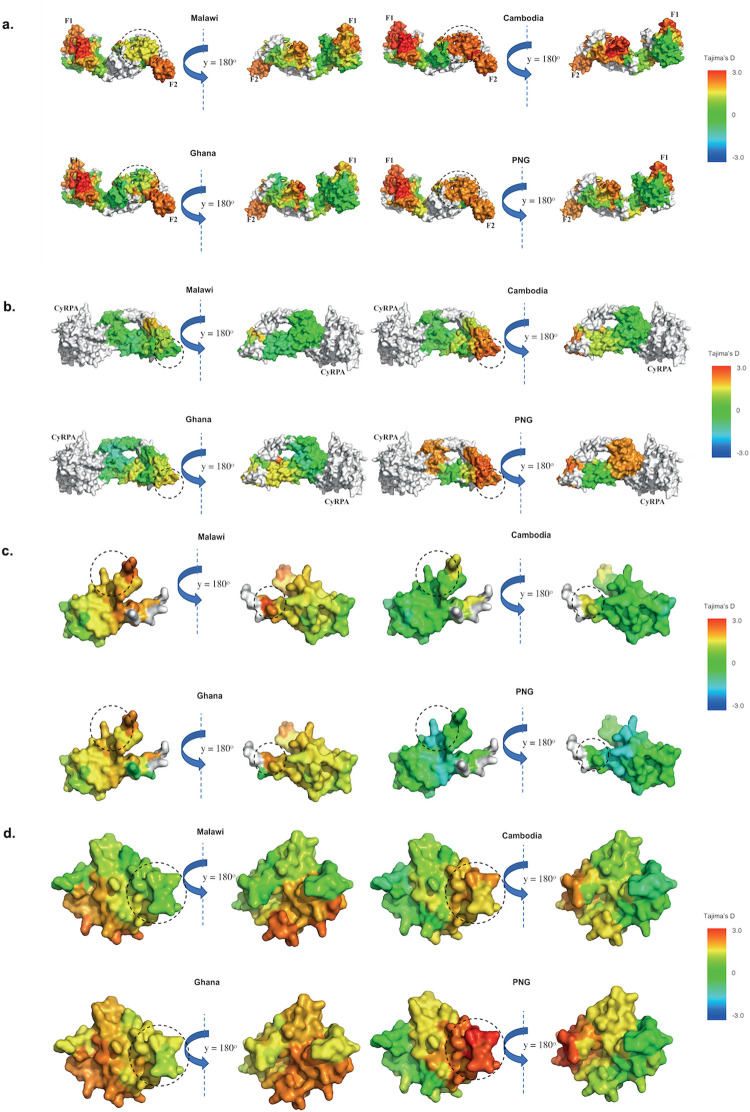
Fig 6. Antigens displaying geographically variable balancing selection hotspots. Antigens were not normalised based on their sizes.
a. Spatially derived Tajima’s D (D*) for EBA175 with incorporation of protein structural information using 15 Å window. 3D7-based ModPipe model of EBA175 (RII) based on 1ZRO template was used. Structure was coloured according to D* scores mapped to each residue with undefined D* were shown in white. The highlighted region (in circle) shows different D* scores between Asia-Pacific and African countries. Sample sizes: Malawi (n = 136), Ghana (n = 237), Cambodia (n = 432), and PNG (n = 112). b. Spatially derived Tajima’s D (D*) calculation for countries from Asia-Pacific and Africa for RH5 with incorporation of protein structural information using 15Å window. Cryo-EM structure of RH5-CyRPA complex (PDB code: 6MPV.B) was used. Structure was coloured according to D* scores mapped to each residue with undefined D* and CyRPA were shown in white. The circle indicates Basigin-binding sites where different D* scores were observed between Asia-Pacific and African countries. Sample sizes: Malawi (n = 142), Ghana (n = 249), Cambodia (n = 433), and PNG (n = 112). c. Spatially derived Tajima’s D (D*) calculations for populations from Asia-Pacific and African regions for CSP (C-terminal) with incorporation of protein structural information using 15 Å window. The crystal structure of the thrombospondin receptor (TSR) domain of CSP [58] (PDB code: 3VDJ, chain A, AA residues: Y306—H376, 3D7 sequence), which consists of Th2R and Th3R (T-cell epitopes) was used [59]. Structure was coloured according to D* scores mapped to each residue with undefined D* were shown in white. The highlighted region (in circle) shows different D* scores between Asia-Pacific and African countries. Sample sizes: Malawi (n = 135), Ghana (n = 223), Cambodia (n = 431), and PNG (n = 111). d. Tajima’s D (D*) calculation for geographic area or countries from the Asia-Pacific and African regions for MSP119 with incorporation of protein structural information using 15 Å window. The structured region of MSP1 (MSP1-19, AA residue: N1607—S1699) is based on the ModPipe homology model using template (PDB code: 1OB1) [60]. The structure was coloured according to D* scores mapped to each residue with undefined D* were shown in white. The circle indicates variable balancing selection hotspots between Asia-Pacific and African countries. Sample sizes: Malawi (n = 101), Ghana (n = 183), Cambodia (n = 270), and PNG (n = 72).