FIGURE 7.
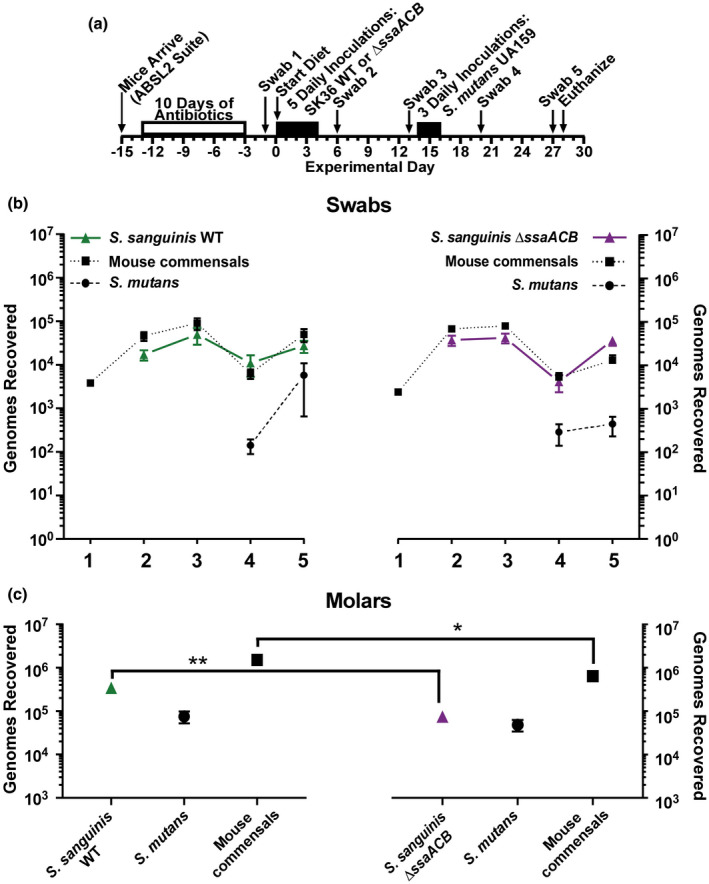
Comparison of oral colonization by Streptococcus sanguinis SK36 WT and ΔssaACB strains, and in subsequent competition with S. mutans UA159 in vivo. (a) Timeline of key events in the experiment. Colonization results for each indicated inoculated S. sanguinis strain (triangles, solid lines), S. mutans UA159 (circles, dashed lines) and mouse oral commensals (squares, dotted lines) from oral swabs 1–5 (b) and from sonicates of mandibular molars (c). S. sanguinis SK36 WT is green (left) and the ΔssaACB mutant is purple (right). Means and standard error (n = 12 mice per cohort) of recovered genomes estimated by qPCR are shown. All samples of each species or group (for commensals) were compared using one‐way ANOVA. Bonferroni's multiple comparisons test was then applied to compare the samples from the WT group to their counterparts in the ΔssaACB group collected at the same time point (*p < .05, **p < .01). There were no significant differences among the swab samples (p > .05). Mice were fed a high‐sucrose powdered diet with sterile drinking water
