Figure 1.
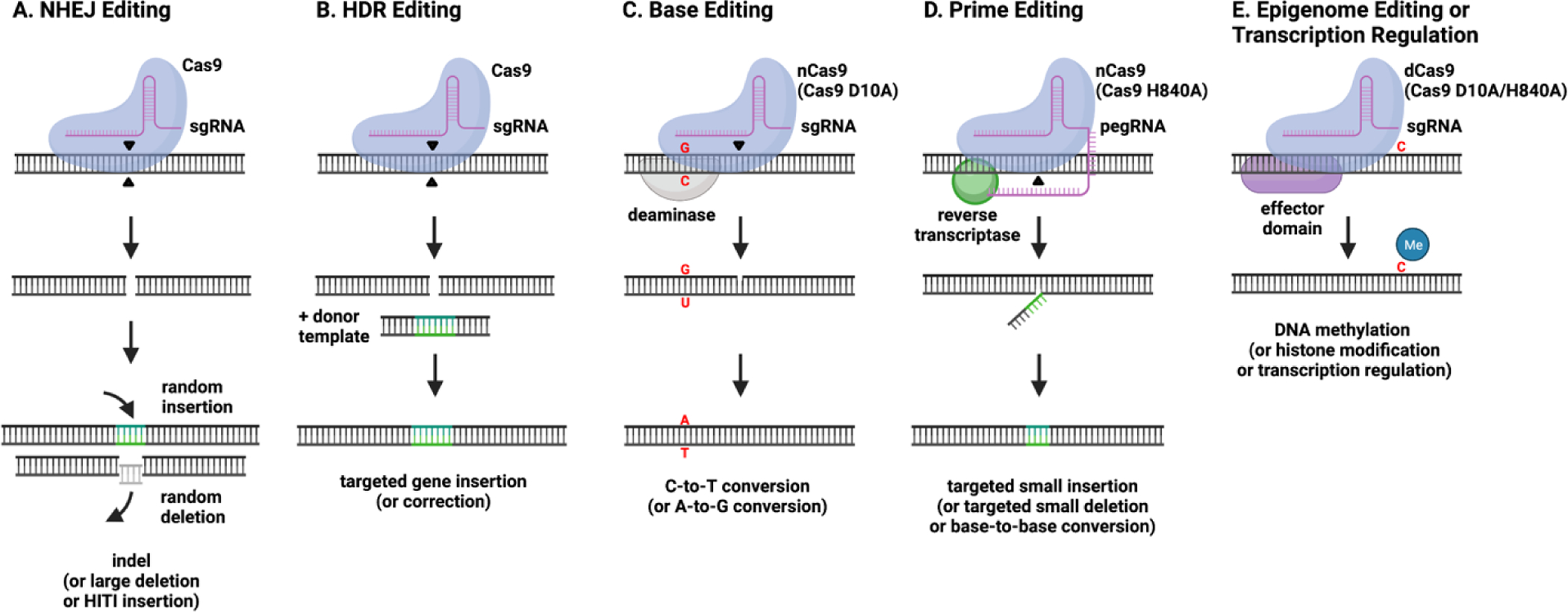
Overview of genome and epigenome editing strategies. A) Non-homologous end joining (NHEJ) is an error prone repair pathway that leads to small insertions or deletions (indels) at the site of Cas9-induced double strand breaks (DSB). When Cas9 is targeted to two genomic sites, large deletions of the intermediate sequence can be generated. Alternatively, targeted insertions can be completed with the addition of a homology-independent targeted integration (HITI) donor template. B) Homology-directed repair (HDR) is a precise DSB repair pathway that utilizes an exogenous DNA donor template with homology arms to achieve targeted gene insertions or corrections. C) Base editing takes advantage of deaminase enzymes to induce C-to-T (cytidine deaminase) or A-to-G (adenosine deaminase) base conversions in a five-base activity window. Cas9 nickase (nCas9; Cas9 D10A) is used to nick a single DNA strand opposite the targeted base in order to facilitate efficient repair. D) Prime editing can achieve targeted insertions, deletions, or base-to-base conversions by the introduction of an extended sgRNA known as a prime editing gRNA (pegRNA) and nCas9 (Cas9 H840A) fused to a reverse transcriptase. E) Epigenome editing and transcription regulation are achieved by guiding effector domains to a targeted DNA site via catalytically inactive dead Cas9 (dCas9; Cas9 D10A/H840A). Effector domains include epigenetic modifying domains, which can alter DNA methylation or histone acetylation, or gene regulatory domains, which directly influence gene expression through interactions with transcriptional machinery.
