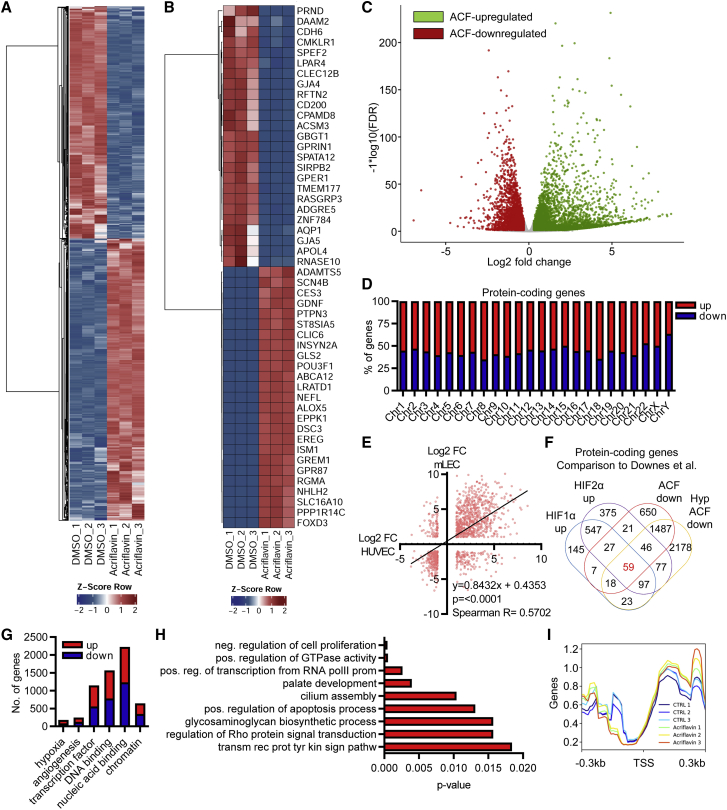
Figure 1.
ACF strongly changes gene expression
(A) Heatmap of RNA-seq after treatment of HUVECs with ACF (10 μM, 16 h) showing significantly (p-value adjusted (padj) < 0.05, log2 fold change (log2FC) >±0.3) altered protein-coding genes. DMSO served as a negative control. 3 replicates are shown. The Z score represents upregulation (red, positive value) or downregulation (blue, negative values) of genes. (B) Heatmap as in (A). The top 25 up- and downregulated protein-coding genes are shown. (C) Volcano plot of RNA-seq after treatment of HUVECs with ACF (10 μM, 16 h), showing significantly altered protein-coding genes. Colored dots indicate significantly differentially expressed genes; padj (false discovery rate [FDR]) < 0.05, log2FC>±0.3). Red, ACF downregulated genes; green, ACF upregulated genes; gray, non-regulated genes (log2FC <±0.3). (D) Chromosomal distribution and percentage of protein-coding genes up- or downregulated with ACF. (E) Correlation analysis (linear regression) of protein-coding genes up- or downregulated with ACF (logFC > ±1.0) of HUVECs and murine lung endothelial cells (mLECs). (F) Venn diagram showing overlap of protein-coding genes significantly downregulated with ACF under normoxic or hypoxic (Hyp) conditions (this study) with genes significantly upregulated after HIF1α or HIF2α overexpression from Downes et al.24 in HUVECs. (G) Number of protein-coding genes up- or downregulated with ACF assigned to selected GO terms. (H) GO enrichment analysis with KOBAS2.0, indicating highly significant GO terms for protein-coding genes up- and downregulated with ACF. (I) Overlay analysis with deeptools2 of the reads with the transcription start sites (TSSs) of all genes, including 0.3 kb up- or downstream.