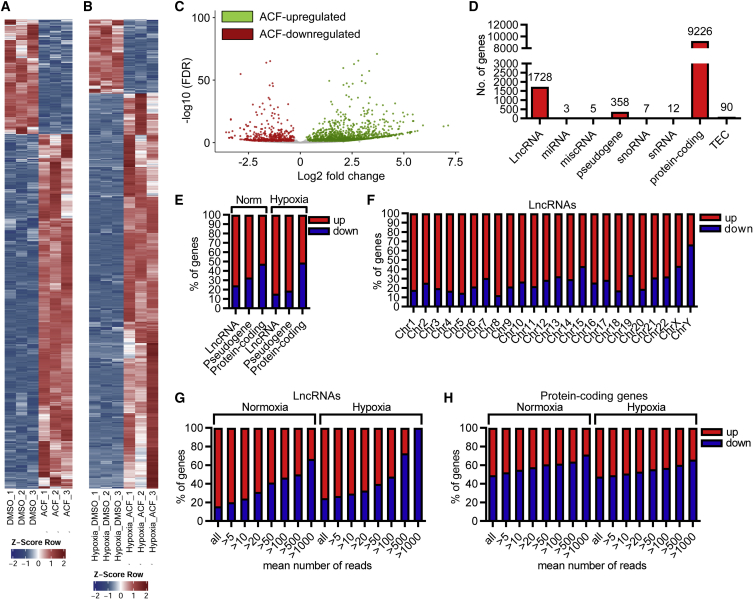
Figure 4.
ACF increases expression of numerous lncRNAs
(A and B) Heatmap of RNA-seq after treatment of HUVECs with ACF (10 μM, 16 h) under normoxic (A) or hypoxic conditions (B), showing significantly (padj <0.05, log2FC >±0.3) altered lncRNAs. DMSO served as a negative CTL. 3 replicates are shown. The Z score represents upregulation (red, positive value) or downregulation (blue, negative values) of genes. (C) Volcano plot of RNA-seq, showing significantly altered lncRNAs under normoxia. Colored dots indicate genes significantly differentially expressed; padj (FDR) < 0.05, log2FC >±0.3. Red, ACF downregulated genes; green, ACF upregulated genes; gray, non-regulated genes (log2FC <±0.3). (D) Numbers of genes from different gene classes significantly altered with ACF under normoxic conditions. (E) Percentage of genes belonging to lncRNAs, pseudogenes, or protein-coding genes up- or downregulated with ACF under normoxic (Norm) or hypoxic conditions. (F) Chromosomal distribution and percentage of lncRNAs up- or downregulated with ACF under normoxic conditions. (G and H) Percentages of lncRNAs (G) and protein-coding genes (H) up- or downregulated by ACF under normoxic or hypoxic conditions, depending on filtering the data for the mean number of reads. All indicates no filter; >5 to >1000 reads indicates that only lncRNAs with read numbers higher than 5–1,000 were used for the analysis.