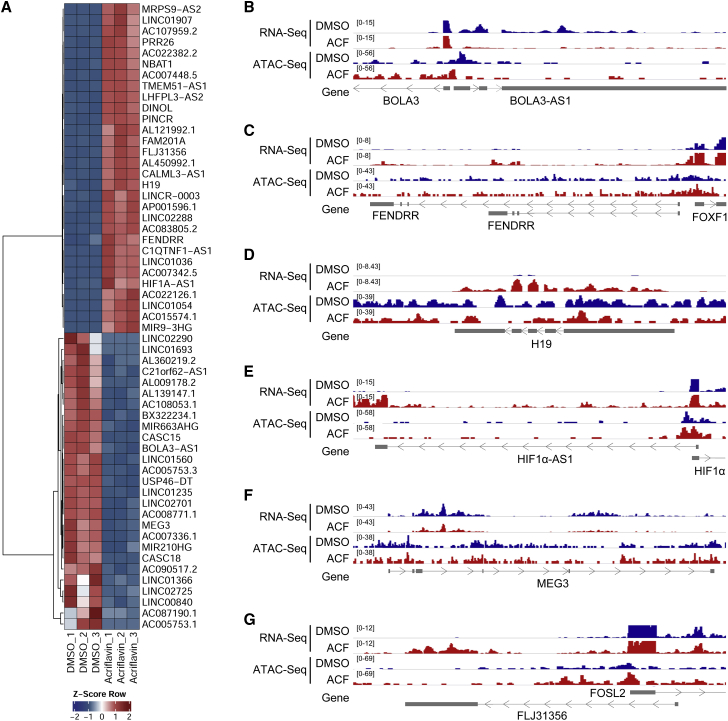
Figure 5.
ACF alters chromatin accessibility on lncRNA loci
(A) Heatmap of RNA-seq of selected significantly altered lncRNAs after treatment of HUVECs with ACF (10 μM, 16 h) under normoxic conditions. DMSO served as a negative CTL. 3 replicates are shown. The Z score represents upregulation (red, positive value) or downregulation (blue, negative values) of genes. (B–G) IGV original traces loaded of RNA-seq and ATAC-seq in HUVECs treated with ACF. Loci for BOLA3-AS1 (B; chr2(hg38):74,146,447–74,152,132), FENDRR (C; chr16(hg38):86,472,458–86,514,791), H19 (D; chr11(hg38):1,993,921–1,998,595), HIF1α-AS1 (E; chr14(hg38):61,679,952–61,697,350), MEG3 (F; chr14(hg38):100,822,031–100,862,947), and FLJ31356 (G; chr2(hg38):28,382,410–28,396,672) are shown. DMSO served as a negative CTL. Numbers in square brackets indicate data range values.