Summary
Epithelial cells lining the oviduct/fallopian tube are essential in reproduction and have been identified as the cell-of-origin in high-grade serous ovarian carcinoma (HGSOC). This protocol describes the generation of organoids from mouse oviduct epithelial cells, providing a powerful in vitro tool to study epithelial homeostasis and malignant transformation. We also outline a protocol for whole-mount immunofluorescence and 3D confocal imaging. In addition, we describe approaches of viral transduction to investigate gene function in organoid development and epithelial cell behavior.
For complete details on the use and execution of this profile, please refer to Ford et al. (2021).
Subject areas: Cell Biology, Cell culture, Cell isolation, Developmental biology, Cancer, Microscopy, Organoids
Graphical abstract
Highlights
-
•
A protocol for the isolation and organoid culture of mouse oviduct epithelial cells
-
•
A procedure for organoid isolation, fixation, and whole mount immunofluorescence
-
•
Organoids retain lineage specific markers and differentiation capacity
-
•
Viral transduction methods to study gene function in disease and homeostasis
Epithelial cells lining the oviduct/fallopian tube are essential in reproduction and have been identified as the cell-of-origin in high-grade serous ovarian carcinoma (HGSOC). This protocol describes the generation of organoids from mouse oviduct epithelial cells, providing a powerful in vitro tool to study epithelial homeostasis and malignant transformation. We also outline a protocol for whole-mount immunofluorescence and 3D confocal imaging. In addition, we describe approaches of viral transduction to investigate gene function in organoid development and epithelial cell behavior.
Before you begin
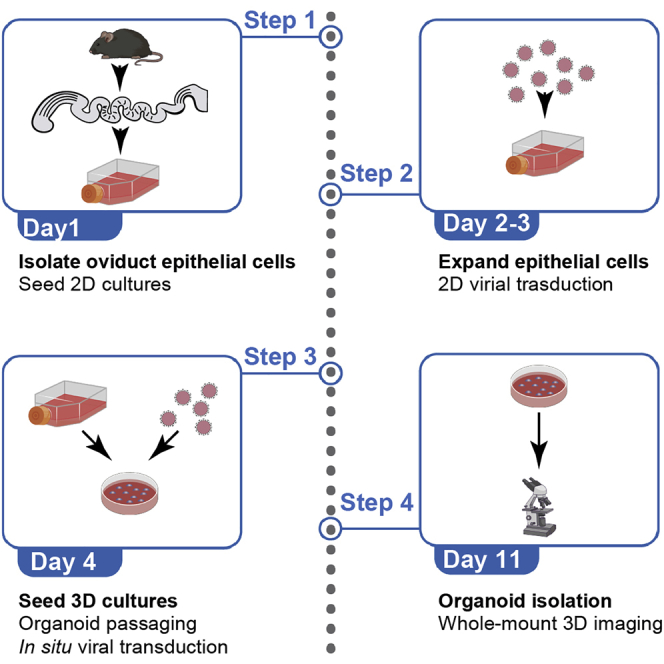
This protocol outlines the procedure to isolate mouse oviduct epithelial cells and propagate organoid cultures. Before attempting this protocol ensure ethical permission for animal experiments has been granted. In this example, we outline a procedure using the entire oviduct. Previous work in our lab has identified distinct distal and proximal epithelial lineages in the mouse oviduct (Ford et al., 2021; Harwalkar et al., 2021). These populations have different characteristics that are maintained in organoid culture. Depending on the experiment, separation of the distal (infundibulum and ampulla) and proximal (isthmus and uterotubal junction) regions may be required. This procedure was optimized using C57BL/6 mice of reproductive age (2–4 months) and includes an intermediate 2D culturing step to expand the epithelial cell population before seeding into 3D cultures. This step overcomes the generally low yields of oviduct epithelial cells during isolation and provides an opportunity for viral transduction. Adaptations for early postnatal stages are also outlined below.
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| PAX8 (1:250) | ProteinTech | Cat# 10336-1-AP; RRID:AB_2236705 |
| Acetylated tubulin (1:250) | Sigma-Aldrich | Cat# T7451; RRID:AB_609894 |
| Sox17 (1:250) | R & D Systems | Cat # AF1924; RRID:AB_355060 |
| Bacterial and virus strains | ||
| Ad-Cre-GFP | University of Iowa, Viral Vector Core | Cat# Iowa-1174 |
| Chemicals, peptides, and recombinant proteins | ||
| 10 x PBS Solution (Phosphate Buffered Saline) | Bio Basic | Cat# PD8117 |
| DMEM/Ham’s F12 | Gibco | Cat# 11320033 |
| HEPES | Gibco | Cat# 15630080 |
| Penicillin-streptomycin | Gibcos | Cat# 15140122 |
| GlutaMAX | Gibco | Cat# 35050-061 |
| Fetal Bovine Serum (FBS) | Wisent Bioproducts | Cat# 090-150 |
| Insulin-transferrin-selenium | Gibco | Cat# 41400045 |
| Human epidermal growth factor (hEGF) | PeproTech | Cat# AF-100-15-500 |
| Bovine pituitary extract | Gibco | Cat# 13028014 |
| Geltrex LDEV-Free Reduced Growth Factor | Gibco | Cat# A1413201 |
| DMEM/F12 advance | Gibco | Cat# LS12634028 |
| B27 | Gibco | Cat# 17504044 |
| TGFBR1 kinase inhibitor IV (SB431542) | BioVision | Cat# 1674-1 |
| Mouse fibroblast growth factor 10 (mFGF10) | PeproTech | Cat# 450-61-25 |
| hNoggin | StemCell Technologies | Cat# 78060.1 |
| hR-spondin1 | StemCell Technologies | Cat# 78213.1 |
| Mouse Wnt family member 3A (mWNT3A) | PeproTech | Cat# 315-20 |
| ROCK inhibitor (Y-27632) | Sigma-Aldrich | Cat# Y0503 |
| Selective γ-secretase inhibitor (Y0-01027) DBZ | Calbiochem | Cat# 209984-56-5 |
| Propyl gallate | Sigma-Aldrich | Cat# 48710 |
| Glycerol | Sigma-Aldrich | Cat# G5516 |
| Formaldehyde 10% | Polysciences | Cat# 04018-1 |
| Collagenase B | Sigma-Aldrich | Cat# COLLB-RO |
| DNAse I (1 U/μL) | Thermo Scientific | Cat# EN0521 |
| Trypsin-EDTA (0.25%), phenol red | Gibco | Cat# 25200056 |
| Bovine Serum Albumin (BSA) | Sigma-Aldrich | Cat# A7030 |
| Tween-20 | Sigma-Aldrich | Cat# P9416 |
| Triton X-100 | Sigma-Aldrich | Cat# X100 |
| Experimental models: Organisms/strains | ||
| Tdtomatoflox/flox (Ai14) female mice C57BL/6 (2–4 months) | JAX | Cat# 007914 |
| Other | ||
| 24-well tissue culture plates | Corning, Falcon | Cat# 353504 |
| 96-well tissue culture plates | Corning, Falcon | Cat# 353072 |
| 0.12 mm Spacer, 8 well, 9 mm | Invitrogen, Secure-Seal | Cat# S24737 |
| Coverslips #1.5 60 × 24 mm | VWR | Cat# CA48393-252-1 |
Materials and equipment
Growth factor and inhibitor stocks
| Reagent | Final stock concentration | Amount | Volume |
|---|---|---|---|
| TGFBR1 kinase inhibitor IV (SB431542) | 10 mM (20,000×) | 5 mg | 1.189 mL DMSO |
| Mouse fibroblast growth factor 10 (mFGF10) | 100 μg/mL (1,000×) | 25 μg | 250 μL sterile PBS + 0.1% BSA |
| Human epidermal Growth Factor (hEGF) | 500 μg/mL (5,000×) | 500 μg | 1 mL sterile PBS + 0.1% BSA |
| Human Noggin | 200 μg/mL (2,000×) | 1000 μg | 5 mL sterile PBS + 0.1% BSA |
| Human R-spondin1 | 600 μg/mL (1,000×) | 500 μg | 833 μL sterile PBS + 0.1% BSA |
| Mouse Wnt family member 3A (mWnt3a) | 100 μg/mL (1,000×) | 10 μg | 100 μL sterile PBS + 0.1% BSA |
| ROCK inhibitor (Y-27632) | 10 mM (1,000×) | 1 mg | 312 μL sterile PBS + 0.1% BSA |
Due to the short half-life of these growth factors and inhibitors, avoid freeze-thaw cycles and follow manufacturer’s recommendations for storage. We recommend to aliquot so that freeze thaw cycles are kept below 5, to avoid degradation and maintain experimental consistency.
Alternatives: Our choice of mouse versus human growth factors is based on previous publications of organoid culture in other systems. While we have not tested different combinations, it is likely these are interchangeable.
2D culture media
| Reagent | Final concentration | Amount |
|---|---|---|
| DMEM/Ham’s F12 | 1× | 91 mL |
| HEPES (1M) | 10 mM | 1 mL |
| Penicillin-Streptomycin (10,000U/mL) | 100 U/mL | 1 mL |
| GlutaMAX (100×) | 1× | 1 mL |
| Insulin-Transferrin-Selenium (100×) | 1× | 1 mL |
| FBS (100 %) | 5 % | 5 mL |
| hEGF (500 μg/mL) | 25 ng/mL | 5 μL |
| Bovine pituitary extract (159 mg/mL | 30 μg/mL | 18.9 μL |
| Total | n/a | 100 mL |
2D media can be stored in 10 mL aliquots at −80°C. Once thawed, store at 4°C and use within 1–2 weeks.
Organoid culture media
| Reagent | Final concentration | Amount |
|---|---|---|
| DMEM/F12 advance | 1× | 47 mL |
| HEPES (1 M) | 10 mM | 500 μL |
| Penicillin-Streptomycin (10,000 U/mL) | 100 U/mL | 500 μL |
| GlutaMAX (100×) | 1× | 500 μL |
| B27 (50×) | 1× | 1 mL |
| SB431542 (10 mM) | 0.5 μΜ | 2.5 μL |
| mFGF10 (100 μg/mL) | 100 ng/mL | 50 μL |
| hEGF (500 μg/mL) | 0.1 μg/mL | 10 μL |
| hNoggin (200 μg/mL) | 100 ng/mL | 25 μL |
| hR-spondin1 (600 μg/mL) | 0.6 μg/mL | 50 μL |
| mWNT3A (100 μg/mL) | 100 ng/mL | 50 μL |
| Total | n/a | 50 mL |
Organoid media can be stored in 1 mL aliquots at −80°C. Once thawed, use immediately and do not refreeze.
CRITICAL: Due to the short half-life of organoid media growth factors and inhibitors it is best to make in small batches and avoid freeze-thaw cycles.
Dissection solution
| Reagent | Final concentration | Amount |
|---|---|---|
| PBS | 1× | 485 mL |
| FBS (100 %) | 2 % | 10 mL |
| Penicillin-Streptomycin (10,000 U/mL) | 100 U/mL | 5 mL |
| Total | n/a | 500 mL |
Dissection solution can be stored at 4°C for up to 1 month.
Collagenase solution
| Reagent | Final concentration | Amount |
|---|---|---|
| Dissection solution | 1× | 940 mL |
| Collagenase B (100 mg/mL) | 5 mg/mL | 50 μL |
| DNase I (1 U/μL) | 0.01 U/μL | 10 μL |
| Total | n/a | 1 mL |
Collagenase solution should be used on the same day and kept on ice.
Alternatives: It is also possible to use collagenase A at the same concentration.
Organoid wash buffer
| Reagent | Final concentration | Amount |
|---|---|---|
| 1× PBS | 1× | 99.9 mL |
| Triton X-100 (100 %) | 0.1 % | 100 μL |
| BSA | 0.2 % | 0.2 g |
| Total | n/a | 100 mL |
Organoid wash buffer can be stored at 4°C for up to 1 month.
Organoid mounting media
| Reagent | Final concentration | Amount |
|---|---|---|
| 10× PBS | 1× | 10 mL |
| Glycerol | 90 % | 89 mL |
| 20 % n-propyl gallate (DMSO) | 0.2 % | 1 mL |
| Total | n/a | 100 mL |
Prepare a stock of 20 % n-propyl gallate by dissolving 1 g in 5 mL DMSO prior to making the mounting media. 1 mL of this solution should be slowly added to the thoroughly mixed PBS:Glycerol (1:9) solution dropwise, while simultaneously stirring rapidly to obtain a final concentration of 0.2%. Mounting media can be stored at 4°C in the dark for several months.
Geltrex
-
•
Thaw a bottle on ice in a cold room/refrigerator (4°C) for between 16–20 h.
-
•
Transfer into a tissue culture hood and mix the Geltrex gently by pipetting using prechilled tips.
-
•
Aliquot 1 mL into prechilled Eppendorf tubes and store at −80°C.
CRITICAL: Ensure Geltrex is kept on ice at all times and mix thoroughly before aliquoting and plating.
CRITICAL: Geltrex is a soluble form of basement membrane extracted from murine Engelbreth-Holm-Swarm (EHS) tumors which can have batch-to-batch variability. It is therefore recommended to perform experiments with a single batch and if required, a comparative proliferation assay when using a new batch for the first time.
Alternatives: This protocol has been optimized with GeltrexTM LDEV-Free Reduced Growth Factor Basement Membrane Matrix. However, several alternatives such as MatrigelTM Growth Factor Reduced (GFR) Basement Membrane Matrix may also be suitable.
Step-by-step method details
Isolation of mouse oviduct epithelial cells and 2D cultures
Timing: 1–2 h
In this section, we outline the steps required to isolate oviduct epithelial cells and seed 2D cultures using the whole oviducts from 5 mice (10 oviducts). Five mice are sufficient to set up approximately 10 wells of a 96-well plate. Single mouse cultures are also possible.
-
1.Dissection of oviducts:
-
a.Prepare collagenase solution as outlined above. Place trypsin-EDTA (0.25%), PBS and 2D culture media in a 37°C water bath.
-
b.Euthanize mice by CO2 administration and/or cervical dislocation.
-
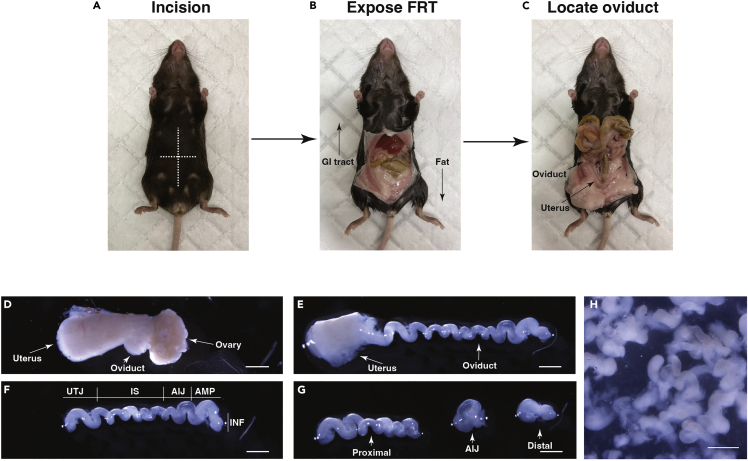
c.Spray the ventral surface of the mouse with 70% ethanol and make an incision down and then across the midline to expose the peritoneal cavity (Figure 1A).
-
d.Move the gastrointestinal tract cranially to reveal the uteri (Figure 1B).
-
e.Follow each uterus laterally to locate the ovary and oviduct. Keeping a portion of the uterus, cut out the ovary and oviduct and place it into an ice cold dissection solution (Figure 1C).
-
f.Under a dissection microscope dissect the oviducts, removing ovary, uterus and connective tissue (Figures 1D–1F).
-
a.
Note: Keeping a portion of the uterus will help manipulation during dissection.
Note: If segregating distal and proximal regions, separate infundibulum and ampulla from the isthmus and uterotubal junction, discarding the ampulla-isthmus junction (Figure 1G).
-
2.Singularization of epithelial cells:
-
a.In a 200 μL drop of collagenase solution pull apart the oviducts into 1 mm segments using forceps and transfer the segments and solution into a sterile 1.5 mL tube containing 200 μL collagenase solution (total solution = 400 μL) (Figure 1H).
-
b.Incubate in a 37°C water bath for 40 min, agitating every 5 min by flicking the tube.Alternatives: A heated 37°C shaker can also be used with agitation every 5–10 min by flicking the tube.
-
c.Centrifuge oviducts/cell suspension at 240×g for 5 min on a tabletop microcentrifuge.
-
d.Resuspend in 500 μL of 37°C PBS to wash the tissue/cells and repeat centrifugation.
-
e.Resuspend in 500 μL Trypsin-EDTA (0.5%) and incubate in a 37°C water bath for 5 min.
-
f.Add 500 μL of 2D media and then pass the suspension 10 times through a 18G needle attached to a 1 mL syringe to dislodge and segregate epithelial cells.
-
g.Using a 200 μL tip, pass the cell suspension through a 40 μm cell strainer into a fresh sterile Eppendorf.
 CRITICAL: Pre-wet tips, cell strainer and syringe with 2D media to prevent loss of cells.Note: If preparing oviduct cultures using early postnatal mice (before sexual maturity which occurs around 40 days after birth), skip steps 2c-e to avoid excessive stromal cell contamination.
CRITICAL: Pre-wet tips, cell strainer and syringe with 2D media to prevent loss of cells.Note: If preparing oviduct cultures using early postnatal mice (before sexual maturity which occurs around 40 days after birth), skip steps 2c-e to avoid excessive stromal cell contamination. CRITICAL: Be careful not to break apart the oviduct segments excessively when passing through the needle. The aim is to dislodge the epithelial cells without breaking the tissue too much, which can result in significant contamination of cultures with stroma cells. See problem 1.
CRITICAL: Be careful not to break apart the oviduct segments excessively when passing through the needle. The aim is to dislodge the epithelial cells without breaking the tissue too much, which can result in significant contamination of cultures with stroma cells. See problem 1.
-
a.
-
3.Seeding 2D cultures:
-
a.Centrifuge cell suspension at 240×g for 5 min.
-
b.Resuspend in 200 μL of 2D media and perform a cell count using a hemocytometer.Note: We recommend counting cells using a hemocytometer, because cell counts using an automated cell counter may be inaccurate due to incomplete singularization of epithelial cells. On average we were able to obtain around 300,000–400,000 cells per 5 mice.
-
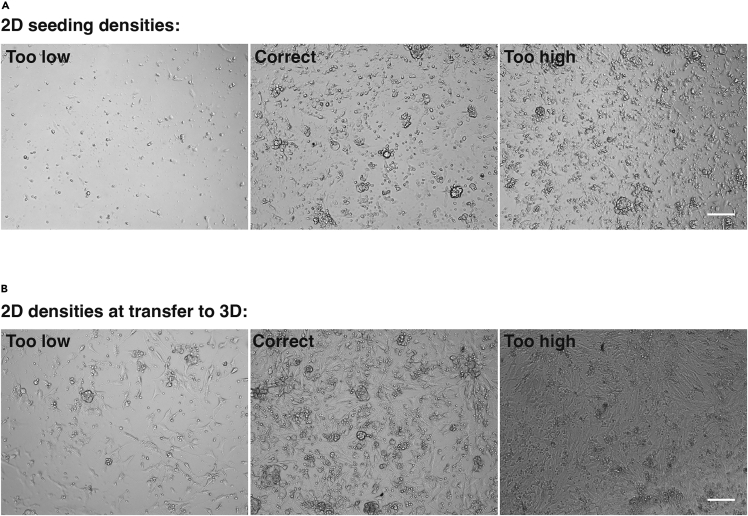
c.Seed cells at a density of 20,000 cells per well in a 96-well flat bottomed tissue culture plate (Corning, Falcon® cat #353072) and fill wells with 200 μL of 2D media (Figure 2A).
-
d.Culture until 75% confluent, usually 2–5 days, in a humidified 37°C incubator supplied with 5% CO2 and change media every 2–3 days (Figure 2B).
 CRITICAL: It is important to seed 2D cultures at a minimum cell density of 20,000 cells per well to ensure successful proliferation. In addition, cells should be transferred to 3D cultures before reaching confluency to capture cells in a proliferating phase. (See problem 2).
CRITICAL: It is important to seed 2D cultures at a minimum cell density of 20,000 cells per well to ensure successful proliferation. In addition, cells should be transferred to 3D cultures before reaching confluency to capture cells in a proliferating phase. (See problem 2).
-
a.
Figure 1.
Dissection of mouse oviducts
(A) Mouse oviducts are located by making a sagittal and transverse incision across the mouse’s abdomen to expose the peritoneal cavity.
(B) The gastrointestinal (GI) tract is then moved cranially and the fat pads caudally to reveal the uteri.
(C) The oviducts can be found adjacent to the ovaries by tracing the uteri laterally.
(D) An isolated oviduct attached to a portion of the uterus and ovary.
(E) A dissected and elongated oviduct after removal of ovary and connective tissue.
(F) After removal of the uterus.
(G) An example of segregation into distal and proximal regions with removal of the ampulla-isthmus junction (AIJ).
(H) Oviducts after being pulled apart into 1 mm fragments for enzymatic digestion. Scale bars = 1 mm. FRT = female reproductive tract, INF = infundibulum, AMP = ampulla, AIJ = ampulla-isthmus junction, IS = isthmus, UTJ = uterotubal junction.
Figure 2.
2D seeding and isolation densities
(A) Brightfield images showing 2D oviduct epithelial cultures after 24 h of culture. To ensure successful expansion of epithelial cells, cultures should be seeded at a density of 20,000 cells per 96-well plate.
(B) Brightfield images showing 2D oviduct epithelial cultures after 3 days of culture. 2D cultures should be transferred to 3D once at 75% confluency, as indicated in the central panel. Scale bars = 100 μm.
Organoid cultures
Timing: 1–2 h
In this section, we outline the steps required to transfer primary 2D oviduct epithelial cultures to organoid cultures.
-
4.Seeding organoid cultures:
-
a.Before beginning, place sterile 24-well tissue culture (Corning, Falcon® cat #353504) plates in a 37°C incubator, thaw Geltrex on ice, and prewarm sterile PBS, trypsin-EDTA (0.25%) and organoid media in a 37°C water bath.
 CRITICAL: Tissue culture plates need to be placed in a 37°C incubator at least 1 h before seeding to ensure Geltrex domes are set in the middle of wells without touching the sides.
CRITICAL: Tissue culture plates need to be placed in a 37°C incubator at least 1 h before seeding to ensure Geltrex domes are set in the middle of wells without touching the sides. -
b.Remove media from 2D cultures and wash wells with 200 μL of PBS.
-
c.To recover cells, add 50 μL of trypsin-EDTA (0.25%) and incubate at 37°C for 10 min, breaking apart cells by pipetting halfway through.
-
d.Add 50 μL of prewarmed 2D media and break apart cells by pipetting before collecting into a sterile Eppendorf, combining cells of the same type.
-
e.Centrifuge the single cell suspension at 240×g for 5 min on a tabletop microcentrifuge followed by resuspension in 200 μL pre-warmed sterile PBS.
-
f.Count cells using a hemocytometer or automated cell counter.
-
g.Add 200 μL of prewarmed sterile PBS to the cell suspension and centrifuge again to wash the cells.
-
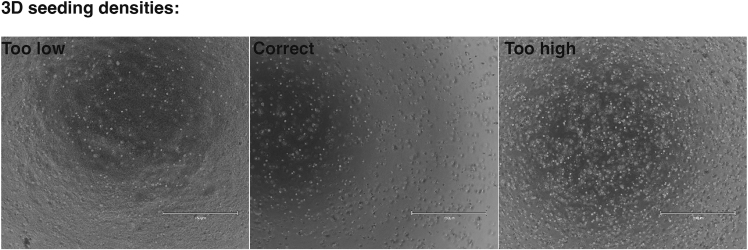
h.Resuspend cells in ice-cold Geltrex at a cell density of 500 cells per μL.
-
i.Seed cells into pre-warmed 24-well tissue culture plates by pipetting a maximum of 50 μL carefully into the center of wells. Carefully flip the plates upside down and place in a 37°C incubator for 30 min to allow the Geltrex to set.
-
j.Once set, add 500 μL of organoid media containing 10 μM Y-27632 and culture in a humidified 37°C incubator supplied with 5% CO2. Fill the remaining wells with 500 μL of sterile PBS to maintain humidity.
-
a.
Figure 3.
3D seeding density
Brightfield images showing oviduct epithelial cells seeded into Geltrex domes. The desired seeding density (500 cells per μL) is indicated in the central panel. If seeded at a lower density (left panel), organoids may fail to form and will have impaired growth. If seeded at a higher density (right panel), organoids will not have enough space to grow and will impede each other’s growth. Scale bar = 750 μm.
-
5.Culturing and passaging organoid cultures:
-
a.Once established, organoid media should be replaced every 2–3 days, without the addition of 10 μM Y-27632. 10 μM Y-27632 should be added only at the time of seeding.
-
b.Organoids should become visible 2–3 days after culture and should be passaged every 7–14 days (Figure 4A).
-
c.To passage organoids, first wash with PBS and add 1 mL of ice-cold PBS to each well. Incubate the plate on ice for 1 h, breaking apart the Geltrex dome halfway through by pipetting.
-
d.After 1 h repeat mixing by pipetting (the Geltrex should now be completely dissolved) and transfer the organoid suspension to a prechilled 15 mL sterile tube and fill up to 10 mL with ice cold PBS, mixing by inversion to ensure the Geltrex has been completely dissolved. See problem 3.
-
e.Centrifuge the organoids at 100×g for 3 min at 4°C.
-
f.Resuspend in 500 mL of prewarmed/37°C trypsin-EDTA (0.25%) and transfer to a sterile 1.5 mL Eppendorf tube. Incubate for 10 min at 37°C, breaking apart organoids at 5 and 10 min by pipetting.
-
g.Centrifuge singularized cells at 240×g for 5 min on a tabletop microcentrifuge.
-
h.Resuspend in 200 μL of prewarmed sterile PBS and count cells using a hemocytometer or automated cell counter.
-
i.Add 200 μL of prewarmed sterile PBS to the cell suspension and centrifuge again to wash the cells.
-
j.Resuspend cells in ice cold Geltrex at a cell density of 500 cells per μL and seed as previously described.
-
a.
-
6.Differentiation of oviduct organoids:
-
a.Oviduct epithelial organoids can be encouraged to undergo multiciliated cell differentiation by Notch inhibition (Ford et al., 2021; Kessler et al., 2015; Xie et al., 2018).
-
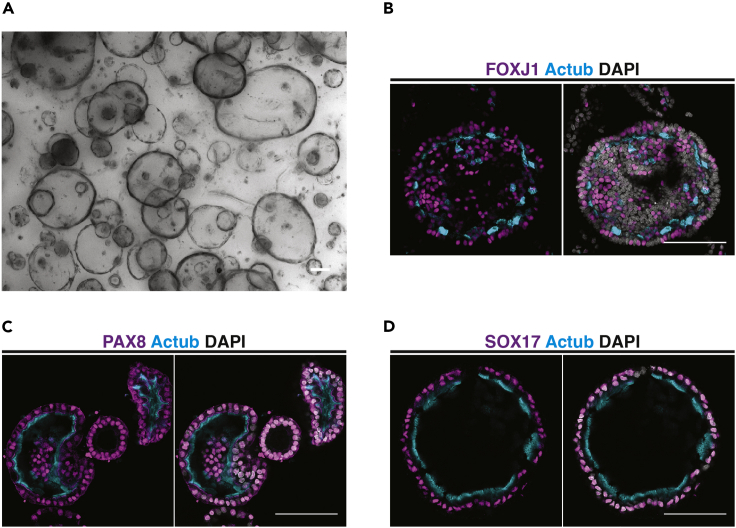
b.To induce differentiation, organoids are allowed to form for 5 days before addition of 1 μM selective y-secretase inhibitor (Y0-01027) and continued culturing for a further 5–7 days (Figures 4B and 4C).
-
a.
Note: Low level spontaneous differentiation into multiciliated cells is also seen after several days in culture.
Figure 4.
Characterization of mouse oviduct organoids
(A) A brightfield image of mouse oviduct epithelial organoids after 7 days in culture.
(B) A whole mount confocal image of a mouse oviduct epithelial organoid after differentiation has been induced by notch inhibition (see step 6a). Multiciliated cells are labeled with FOXJ1 and cilia with Actub (acetylated tubulin). Nuclei are labeled with DAPI.
(C and D) Whole mount imaging of differentiated mouse oviduct epithelial organoids showing retention of oviduct epithelial markers PAX8, SOX17 and cilia labeled with Actub. Nuclei are labeled with DAPI. Scale bars = 100 μm.
Whole-mount immunofluorescence and 3D confocal microscopy
Timing: 3 days
In this section, we outline the procedure to characterize oviduct organoids by whole mount immunofluorescence staining and 3D confocal imaging. This protocol is an adaptation of a previously published general organoid staining protocol (Dekkers et al., 2019).
-
7.Fixation:
-
a.To isolate organoids, first wash with PBS and add 1 mL of ice-cold PBS to each well. Incubate the plate on ice for 1 h, breaking apart the Geltrex dome halfway through by pipetting.
-
b.After 1 h repeat mixing by pipetting, the Geltrex should now be completely dissolved.
-
c.Coat a pre-chilled 15 mL sterile tube with 1% (wt/vol) ice cold PBS-BSA (Bovine Serum Albumin). Also coat pipette tips with the same solution by pipetting up and down 3 times before handling organoids.
 CRITICAL: Until organoids are fully fixed (step i) coat tubes and tips with 1% (wt/vol) ice cold PBS-BSA before contact with organoids to prevent organoids from adhering to plastic surfaces.
CRITICAL: Until organoids are fully fixed (step i) coat tubes and tips with 1% (wt/vol) ice cold PBS-BSA before contact with organoids to prevent organoids from adhering to plastic surfaces. -
d.Transfer the organoid suspension to pre-chilled 15 mL sterile tubes on ice, combining organoids under the same conditions.
-
e.Collect any remaining organoids in the wells with 1 mL 1% (wt/vol) ice cold PBS-BSA.
-
f.Fill up to 10 mL with ice cold PBS, mixing by inversion to ensure the Geltrex has been completely dissolved.
-
g.Centrifuge organoids at 100 g for 3 min at 4°C.
 CRITICAL: Organoids should form a pellet without any remaining Geltrex. Undissolved Geltrex can stick organoids together and impair immunofluorescence by preventing penetration of antibodies or producing high background staining (See problem 3).
CRITICAL: Organoids should form a pellet without any remaining Geltrex. Undissolved Geltrex can stick organoids together and impair immunofluorescence by preventing penetration of antibodies or producing high background staining (See problem 3). -
h.Organoids are then resuspended in 1 mL ice cold 4% (wt/vol) PBS-PFA (Paraformaldehyde) and incubated on ice for 45 min to fix, mixing the organoids halfway through by pipetting.
-
i.Fill up to 10 mL with 0.1% (vol/vol) PBS-Tween20 and mix by inversion before centrifugation at 100×g for 5 min at 4°C.
 Pause point: Organoids can be stored in 0.1% (vol/vol) PBS-Tween20 for several days before starting immunofluorescence staining.
Pause point: Organoids can be stored in 0.1% (vol/vol) PBS-Tween20 for several days before starting immunofluorescence staining.
-
a.
-
8.Immunofluorescence:
-
a.Resuspend organoids in 4°C organoid wash buffer (OWB) (0.1% (v/v), 0.2% (wt/v) PBS-Triton-X-BSA) and segregate into required wells of a 24-well plate, containing at least 200 μL OWB per well before addition of organoids.
-
b.Leave on ice for 15 min for blocking.
-
c.Add 200 μL of OWB to each well and to an empty well as a reference.
-
d.Wait for the organoids to settle to the bottom then remove 200 μL from each well using the empty well as a reference to ensure 200 μL if left in each well.Alternatives: Plates can be placed on a gentle horizontal shaker to bring organoids to the center, allowing for removal of liquids by pipette from the side.
-
e.Add 200 μL of OWB containing a 2× dilution of the primary antibodies resulting in 400 μL of a 1× antibody solution per well and incubate for between 16 to 20 h at 4°C on a horizontal shaker.
-
f.On the next day, add 1 mL of OWB to each well and wait 2 min for the organoids to sink to the bottom.
-
g.Remove 1 mL of OWB and replace it with a fresh 1 mL of OWB.
-
h.Leave on a gentle shaker for 2 h at 21°C.
-
i.Repeat steps 8g and h 2 more times.
-
j.Wait 2 min for organoids to sink to the bottom then remove 1 mL of OWB from each well.
-
k.Add 200 μL of OWB containing a 2× dilution of the secondary antibodies resulting in 400 μL of a 1× antibody solution per well and incubate for between 16 to 20 h at 4°C on a horizontal shaker.
-
l.On the next day, add 1 mL of OWB to each well and wait 2 min for the organoids to sink to the bottom.
-
m.Remove 1 mL of OWB and replace it with a fresh 1 mL of OWB.
-
n.Leave on a gentle shaker for 2 h at 21°C.
-
o.Repeat steps 8m and n 2 more times.
-
p.Wait 2 min for organoids to sink to the bottom then remove 1 mL of OWB from each well.
-
q.Add 200 μL OWB containing a 2× dilution of DAPI and incubate for 10 min.
-
r.Add 1 mL of OWB to each well and wait 2 min for the organoids to sink to the bottom.
-
s.Remove 1 mL of OWB and replace it with 1 mL of PBS.
-
a.
-
9.Mounting:
-
a.Stick a 0.12 mm deep spacer (e.g., Secure-SealTM 8 well, 9 mm) onto a coverslip.
-
b.Pipette organoids into the well created by the spacer and remove as much left-over fluid as possible.
-
c.Add a drop of organoid mounting media to each well containing organoids and seal by sticking another coverslip on top.
 CRITICAL: Addition of the mounting media and second coverslip can dislodge the organoids. Removing as much of the left-over fluid during the transfer and allowing the organoids to dry a little in place can help them stick to the coverslip.
CRITICAL: Addition of the mounting media and second coverslip can dislodge the organoids. Removing as much of the left-over fluid during the transfer and allowing the organoids to dry a little in place can help them stick to the coverslip. -
d.Mounted organoids sandwiched between two coverslips should be stored at 4°C in the dark and imaged as soon as possible (see problem 4).
-
a.
Viral transduction
In this section, we outline steps to perform viral transduction of oviduct organoids in situ and during 2D expansion. Transduction during 2D expansion allows for the generation of clonal organoids within a mixed culture of transduced and non-transduced cells. This can be particularly useful when assaying for effects of gene function on organoid size, which we have shown to be dependent on organoid density (Ford et al., 2021). Separation of transduced cells is also possible using viral constructs containing fluorescent reporters and fluorescence-activated cell sorting (FACS). In situ transduction on the other hand allows for the manipulation of single cells within a fully formed organoid, which can be a good model of cancer initiation. As an example of 2D and in situ transduction we detail protocols using a Cre-expressing adenovirus (Ad-Cre-GFP (a kind gift from Dr. Luke McCaffrey, McGill University)) with an inducible fluorescent reporter mouse line, Tdtomatoflox/flox (Madisen et al., 2010).
-
10.Viral transduction during 2D expansion:
-
a.Isolate oviduct epithelial cells from Tdtomatoflox/flox mice and seed in 2D cultures as outlined in steps 1–3.
-
b.Culture cells until 40%–50% confluency, usually around 2 days.
-
c.Dilute Cre-expressing adenovirus to a final working concentration of 1.2 × 106 pfu/mL in pre-warmed 2D media.Note: The concentration of virus will depend on the desired outcomes of the experiment and the virus used. This should be optimized for each experiment. The concentration in this example produces robust transduction and can be used as a guide (Figure 5A).
-
d.Remove media from 2D cultures and replace it with 200 μL of 2D media containing the adenovirus.
-
e.Culture for between 16 to 20 h in a humidified 37°C incubator supplied with 5% CO2.
-
f.On the next day either replace 2D media and continue culturing to 75% confluency or transfer directly to 3D cultures as outlined in step 4.
-
g.Successful transduction can be confirmed by the expression of Tdtomato after 3–5 days (Figure 5B) (see problem 5).Alternatives: To reduce the amount of virus particles needed for an experiment it is possible to use 50 μL of virus containing media for each well, culture for 4 h and then top up to 200 μL before culturing for between 16 to 20 h.
-
a.
-
11.Viral transduction in situ:
-
a.Oviduct epithelial organoids are generated from Tdtomatoflox/flox mice as outlined in steps 1–4.
-
b.Organoids are allowed to form for 3–5 days before transduction.
-
c.Dilute Cre-expressing adenovirus to a final working concentration of 1.2 × 106 pfu/mL in prewarmed organoid media.
-
d.Remove media from organoid cultures and replace it with organoid media containing the adenovirus.
-
e.Culture for between 16 to 20 h in a humidified 37°C incubator supplied with 5% CO2.
-
f.Replace media with fresh organoid media.
-
g.Successful transduction can be confirmed by the expression of Tdtomato after 3–5 days (Figure 5C).
-
a.
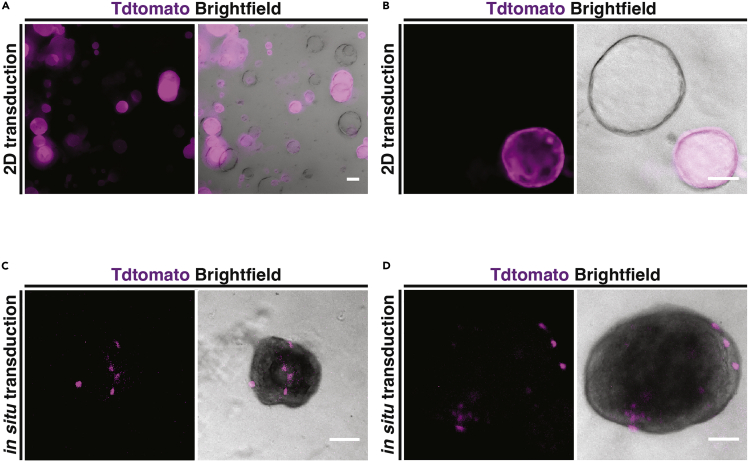
Figure 5.
Viral transduction of mouse oviduct epithelial organoids
(A and B) Live images of mouse oviduct epithelial organoids generated from TdtomatoFlox/Flox mice after transduction during 2D culture with a cre-expressing Adenovirus. Tdtomato expression was identified in some organoids indicating successful viral transduction.
(C and D) Live images of mouse oviduct epithelial organoids generated from TdtomatoFlox/Flox mice after in situ transduction on day 7 of culture with a cre-expressing Adenovirus. Tdtomato expression was identified in a subpopulation of epithelial cells within some organoids, indicating successful viral transduction. Scale bars = 100 μm.
Expected outcomes
In this protocol, we describe a step-by-step guide for the isolation of mouse oviduct epithelial cells and the generation of organoid cultures. These organoids retain the molecular markers and differentiation capacity of oviduct epithelial cells in addition to their regional identities (Ford et al., 2021). We also detail techniques to perform whole mount immunofluorescence and viral transduction. Organoids provide a versatile tool to study the molecular mechanisms of epithelial homeostasis and malignant transformation. The oviduct epithelium is essential for reproduction, interacting with and aiding the transport of gametes and embryos (Li and Winuthayanon, 2017). In addition, it is the cell-of-origin of HGSOC, the most common and deadly subtype of ovarian cancer (Kroeger and Drapkin, 2017; Labidi-Galy et al., 2017). The majority of HGSOCs are detected late due to asymptomatic development which has hindered our understanding of early cancer development. Mouse oviduct organoids represent a valuable tool to study these events in vitro and due to the extensive catalog of genetically engineered mouse models available, it provides a complementary model to human fallopian tube organoid cultures.
Limitations
The above protocol generates mouse oviduct organoids using crude isolation of epithelial cells. Using this technique, it is impossible to generate pure epithelial cultures without some level of stroma cell contamination. In our experience however, all organoids expressed markers of oviduct epithelial cells (PAX8, FOXJ1, SOX17) and little contamination was seen in cultures, likely due to the incomplete digestion of the oviducts and selection of epithelial cells during culturing. If pure cultures are required, it is possible to use our protocol with FACS to remove the stromal compartment. This can be performed either during epithelial cell isolation or after 2D expansion. However, cell yields using FACS are significantly lower and require the use of significantly more mice per experiment.
Generation of mouse oviduct organoids with the described protocol uses an intermediary step of 2D culture. We include this step to expand the epithelial population thereby significantly reducing the number of mice required for each experiment. However, it is becoming clearer that 2D culture techniques significantly alter a cell’s behavior compared to the in vivo condition (Jensen and Teng, 2020). While oviduct organoids generated using our protocol retain expression of key markers, differentiation capacity and lineage identity, changes and selection induced by 2D culturing should be considered. During the optimization of this protocol, we attempted direct seeding of epithelial cells into organoid cultures. We found however that these cultures tended to be less consistent and contained significant stromal contamination.
Troubleshooting
Problem 1
Significant stromal contamination in organoid cultures (see step 2).
Potential solution
Using our crude epithelial cell isolation protocol, it is impossible to remove all stromal cells. The small number of stromal cells that remain in general do not interfere with organoid growth and may in fact release factors that promote epithelial cell proliferation and survival. In some cases, however, significant stromal contamination can occur which can result in organoid cultures becoming overgrown with stromal cells. To reduce stromal contamination, ensure all connective tissue is removed during dissection and decrease the number and/or force at which oviducts are passed through the needle during isolation. Omitting the trypsinization step can also reduce the amount of stromal contamination but will impact cell yield. To remove stromal cells from 3D cultures, carefully take out the Geltrex dome without dislodging the stromal cells growing on the bottom of the plate and continue the recovery of organoids in a new well.
Problem 2
Organoids fail to grow after successful 2D culture (see step 4).
Potential solution
Failure for organoids to form after 2D culturing is generally either a result of incorrect seeding density, overgrown 2D cultures or significant stromal contamination (see problem 1). Ensure cells are transferred from 2D to 3D before reaching confluency to capture cells in a proliferation phase and seed at 500 cells per μL. See Figures 2 and 3 for examples of correct densities.
Problem 3
Incomplete removal of Geltrex during organoid isolation (see step 7).
Potential solution
If incubation in PBS on ice is insufficient to isolate organoids from the Geltrex, specialized recovery solutions such as CorningTM Cell Recovery Solution can help to remove the basement membrane.
Problem 4
Organoids appear deformed/folded (see step 9).
Potential solution
In some cases, large organoids can collapse and become folded during fixation and processing for imaging. If this occurs, we suggest optimizing the fixation and try different fixatives such as formalin or 2% (vol/vol) PFA-glutaraldehyde.
Problem 5
Heterogeneous organoids (see step 9).
Potential solution
2D viral transduction using our protocol generally produces organoids which are homogeneous for viral transduction (Tdtomato +) or wild type, indicating that organoids are likely coming from a single cell. However, in some cases, we have identified heterogeneous organoids. This could be due to organoids forming from a small group of epithelial cells due to incomplete singularization or seeding at too higher density. These steps should be carefully optimized in addition to titration of viral load to promote the formation of homogeneous organoids. Alternativity FACS can also be used to isolate transduced cells and seed separate cultures.
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Dr. Yojiro Yamanka (Yojiro.yamanaka@mcgill.ca).
Materials availability
This study did not generate new unique reagents.
Acknowledgments
We thank the McGill Advanced Bioimaging Facility (ABIF) and Comparative Medicine and Animal Research Centre (CMARC). This work was supported by the Canadian Cancer Society (CCS) innovation grants (Haladner Memorial Foundation no. 704793: no. 705874), a CCS i2I grant (no. 706320), and a Cancer Research Society operation grant (no. 23237). M.J.F. was supported by Canderel, CRRD, and FRQS postdoc fellowships. K.H. was supported by CRRD and Alexander McFee (Faculty of Medicine) and Rolande and Marcel Gosselin studentships.
Author contributions
Y.Y. conceived the study. M.J.F. optimized the protocol, performed most of the experiments, and prepared the manuscript. K.H. helped in the optimization of the protocol and provided some of the data. K.H., Y.Y., and M.J.F. edited the manuscript.
Declaration of interests
The authors declare no competing interests.
Contributor Information
Matthew J. Ford, Email: matthew.ford@mail.mcgill.ca.
Yojiro Yamanaka, Email: yojiro.yamanaka@mcgill.ca.
Data and code availability
This study did not generate/analyze datasets/code.
References
- Dekkers J.F., Alieva M., Wellens L.M., Ariese H.C.R., Jamieson P.R., Vonk A.M., Amatngalim G.D., Hu H., Oost K.C., Snippert H.J., Visvader J.E. High-resolution 3D imaging of fixed and cleared organoids. Nat. Protoc. 2019;14 doi: 10.1038/s41596-019-0160-8. [DOI] [PubMed] [Google Scholar]
- Ford M.J., Harwalkar K., Pacis A.S., Maunsell H., Wang Y.C., Badescu D., Teng K., Yamanaka N., Bouchard M., Ragoussis J., Yamanaka Y. Oviduct epithelial cells constitute two developmentally distinct lineages that are spatially separated along the distal-proximal axis ll ll Oviduct epithelial cells constitute two developmentally distinct lineages that are spatially separated along the di. CellReports. 2021;36:109677. doi: 10.1016/j.celrep.2021.109677. [DOI] [PubMed] [Google Scholar]
- Harwalkar K., Ford M.J., Teng K., Yamanaka N., Yang B., Burtscher I., Lickert H., Yamanaka Y. Anatomical and cellular heterogeneity in the mouse oviduct-- its potential roles in reproduction and preimplantation development. Biol. Reprod. 2021:1–13. doi: 10.1101/2020.08.23.263772. [DOI] [PubMed] [Google Scholar]
- Jensen C., Teng Y. Is it time to start transitioning from 2D to 3D cell culture. Front. Mol. Biosciences. 2020;7:1–15. doi: 10.3389/fmolb.2020.00033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessler M., Hoffmann K., Brinkmann V., Thieck O., Jackisch S., Toelle B., Berger H., Mollenkopf H.J., Mangler M., Sehouli J., Fotopoulou C. The Notch and Wnt pathways regulate stemness and differentiation in human fallopian tube organoids. Nat. Commun. 2015;8989 doi: 10.1038/ncomms9989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroeger P., Drapkin R. Pathogenesis and heterogeneity of ovarian cancer. Current Opinion in Obstetrics and Gynecology. 2017;29:26–34. doi: 10.1097/GCO.0000000000000340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labidi-Galy I., Papp E., Hallberg D., Niknafs N., Adleff V., Noe M., Bhattacharya R., Novak M., Jones S., Phallen J., et al. High grade serous ovarian carcinomas originate in the fallopian tube. Nature Communications. 2017;8:1093. doi: 10.1038/s41467-017-00962-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S., Winuthayanon W. Oviduct : roles in fertilization and early embryo development. Journal of Endocrinology. 2017;232:R1–R26. doi: 10.1530/JOE-16-0302. [DOI] [PubMed] [Google Scholar]
- Madisen L., Zwingman T.A., Sunkin S.M., Oh S.W., Zariwala H.A., Gu H., Ng L.L., Palmiter R.D., Hawrylycz M.J., Jones A.R., Lein E.S. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat. Neurosci. 2010;13 doi: 10.1038/nn.2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Y., Park E.S., Xiang D., Li Z. Long-term organoid culture reveals enrichment of organoid-forming epithelial cells in the fimbrial portion of mouse fallopian tube. Stem Cell Res. 2018;32:51–60. doi: 10.1016/j.scr.2018.08.021. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
This study did not generate/analyze datasets/code.