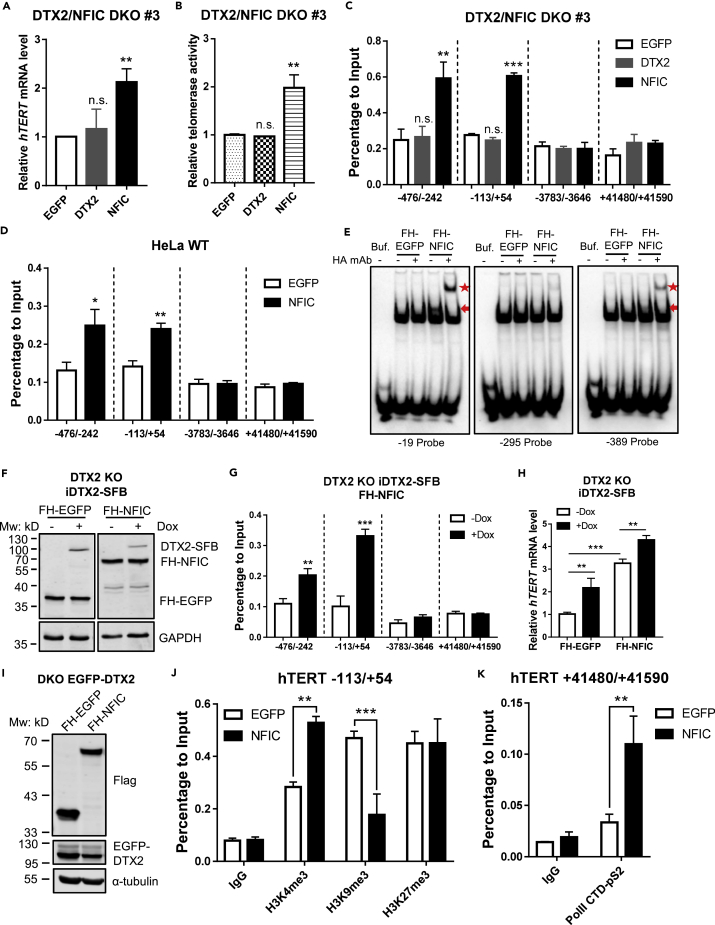
Figure 6.
DTX2 promotes NFIC association with the hTERT promoter for hTERT transcriptional activation
(A and B) FH-tagged EGFP, DTX2, or NFIC was ectopically expressed in the DTX2/NFIC double knockout (DKO) line #3. Relative hTERT mRNA level (A) and telomerase activity (B) were assessed. Data were shown as mean ± SD (N = 3). ∗∗p< 0.01; n.s., not significant.
(C) Cells from (A) were also examined by ChIP-qPCR with an anti-HA antibody and primers for the indicated region (relative to hTERT TSS). Enrichment of the examined regions in ChIP samples was normalized to input. Data were shown as mean ± SD (N = 3). ∗∗p< 0.01; ∗∗∗p< 0.001; n.s., not significant.
(D) HeLa cells stably expressing FH-NFIC were analyzed in ChIP-qPCR with an anti-HA antibody and primers for the indicated region (relative to hTERT TSS). Results were normalized to input. Data were shown as mean ± SD (N = 3). ∗p< 0.05; ∗∗p< 0.01.
(E) In the electrophoretic mobility shift assay (EMSA), cell lysate from HEK293T cells transiently expressing FH-EGFP or FH-NFIC was incubated with biotin-labeled double-stranded DNA probes containing the flanking sequences surrounding regions −19, −295, or −389 upstream of hTERT TSS. An anti-HA antibody was used to super-shift the protein-DNA complex. Reaction mixtures were resolved on a nondenaturing PAGE gel and blotted with streptavidin. Red arrow indicates protein-DNA complex. Red star indicates super-shifted complexes.
(F–H) Dox-inducible DTX2-SFB (iDTX2-SFB) were reintroduced into DTX2 KO cells with FH-EGFP or FH-NFIC expression. Cells were cultured with −/+Dox (1 μg/mL) for 48 h before western blot (F), ChIP-qPCR analysis of indicated regions (relative to hTERT TSS) (G), and RT-qPCR for relative hTERT mRNA level (H). Data were shown as mean ± SD (N = 3). ∗∗p< 0.01; ∗∗∗p< 0.001. For all panels, p values were calculated using Student’s t test.
(I) DTX2/NFIC DKO cells stably expressing FH-tagged EGFP or NFIC were reintroduced with EGFP-DTX2 and examined by western blot using the indicated antibodies.
(J) ChIP-qPCR analysis was done using cells from (I) and anti-H3K4me3, anti-H3K9me3, and anti-H3K27me3 antibodies. Primers span areas within the hTERT core promoter region (−113/+54). Enrichment of specific regions was normalized to input. Data were shown as mean ± SD (N = 2). p values were calculated using Student’s t test. ∗∗p< 0.01; ∗∗∗p< 0.001.
(K) ChIP-qPCR analysis was done using cells from (I) and anti-Pol II-pS2 antibody. Primers span areas in the last exon of hTERT (+41,480/+41,590). Enrichment of specific regions was normalized to input. Data were shown as mean ± SD (N = 2). p values were calculated using Student’s t test. ∗∗p< 0.01. See also Figure S6.