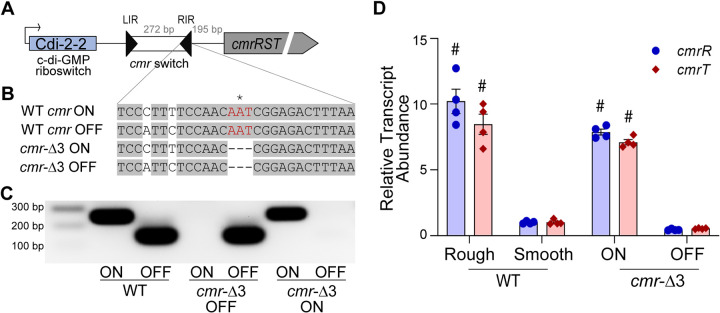
FIG 1.
Generation of genetically phase-locked strains. (A) Diagram of the cmrRST locus. The previously identified c-di-GMP riboswitch Cdi-2-2 and putative associated promoter are indicated. Black triangles represent the inverted repeats flanking the invertible DNA sequence (cmr switch). The OFF orientation corresponds to the sequence present in the R20291 reference genome (GenBank accession number FN545816); the ON orientation corresponds to the inverse of that sequence. (B) Sequence of the right inverted repeat (RIR) of the cmr switch. Gray shading indicates sequence identity between the imperfect inverted repeats of the RIR in each orientation. Three nucleotides (shown in red) were deleted from the RIR to lock the cmr switch in the ON and OFF orientations. The asterisk (*) indicates the nucleotide at the site of recombination. (C) Orientation-specific PCR to detect each orientation of the cmr switch in WT, cmr-Δ3 OFF, and cmr-Δ3 ON strains. (D) qRT-PCR analysis for the cmrR and cmrT transcripts in WT rough and smooth isolates, cmr-Δ3 OFF, and cmr-Δ3 ON. Data from four biological replicates were analyzed using the ΔΔCT method with rpoC as the reference gene and normalization to the WT smooth samples. Shown are means and standard error. #, P < 0.0001; two-way analysis of variance (ANOVA) with Sidak’s multiple-comparison posttest compared to the WT/smooth condition.