FIG 1.
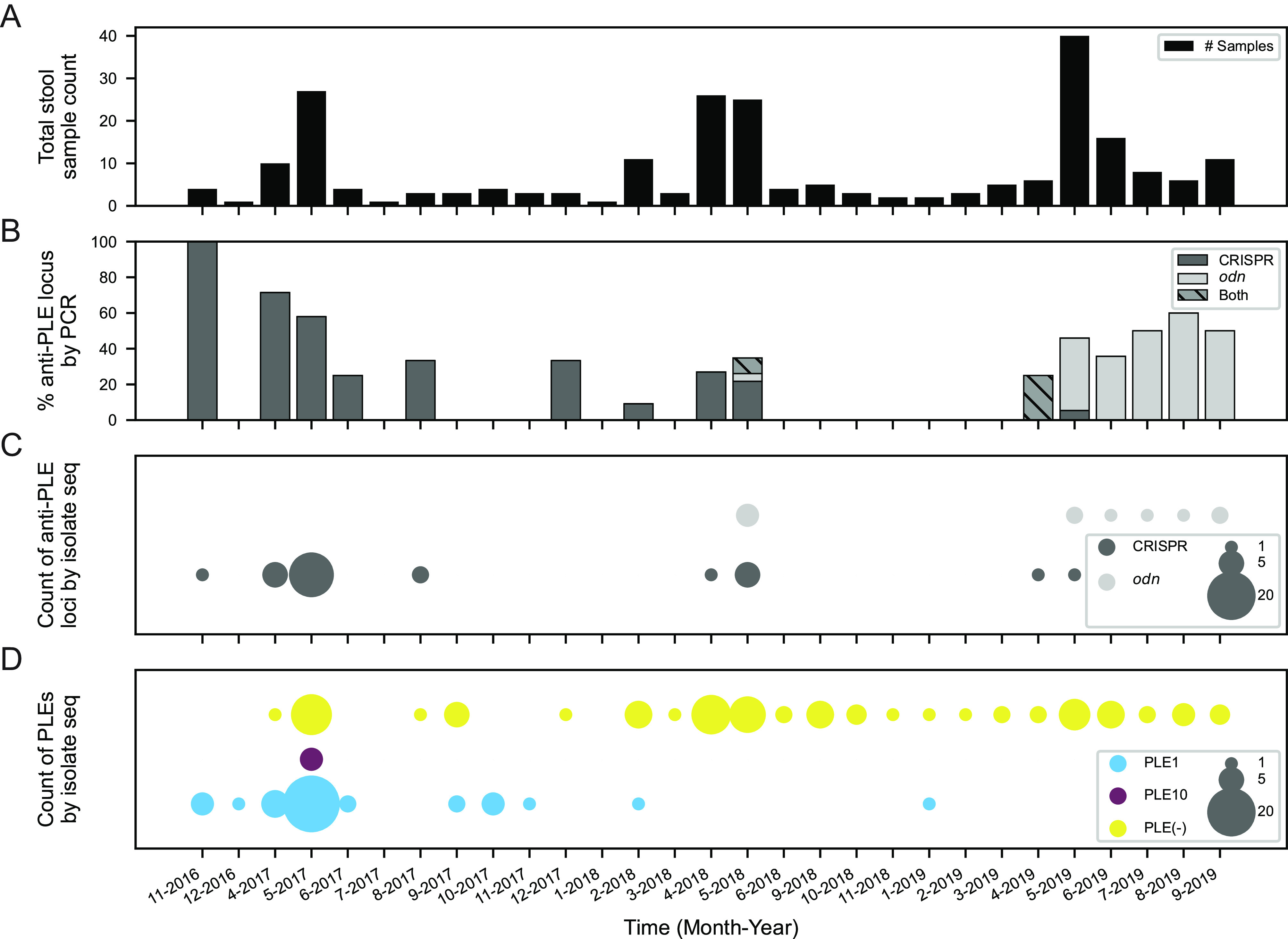
Surveillance of V. cholerae PLE and ICP1 anti-PLE loci in Bangladesh. (A) Total counts of patient stool samples collected and analyzed by month and year of collection (239 total samples) (Data Set S1). (B) Percentage of stool samples in a given month that screened PCR positive for ICP1-encoded anti-PLE counterdefense loci CRISPR-Cas (n = 36), odn (n = 32), or both (n = 3). (C) Number of whole-genome-sequenced ICP1 isolates (44 total) from stool samples over the surveillance period by the type of anti-PLE counterdefense locus CRISPR-Cas or odn. (D) Number of whole-genome-sequenced V. cholerae isolates (148 total) from stool samples over the surveillance period by PLE variant type. PLE(−) indicates no PLE detected.
