FIG 2.
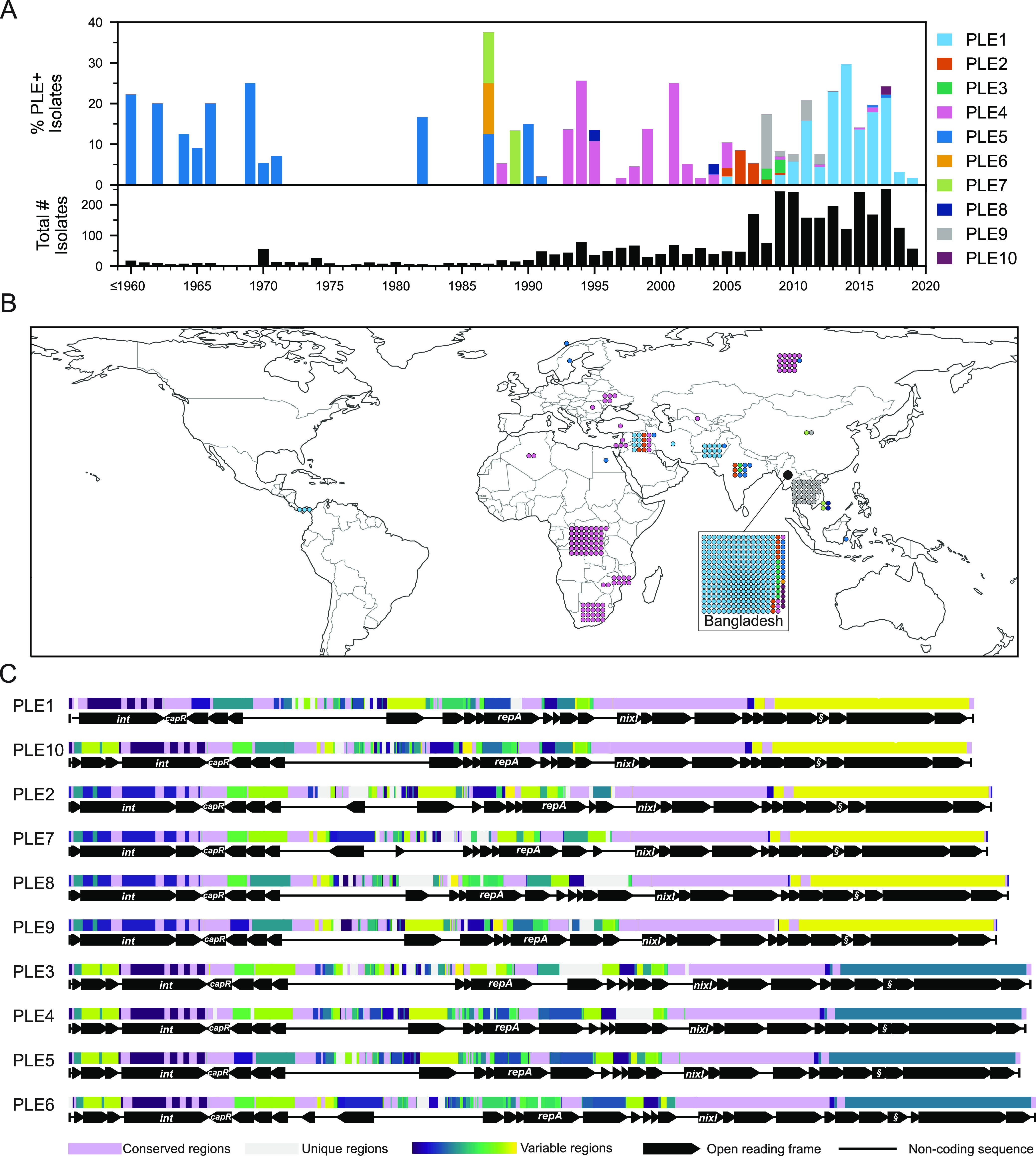
Genetic, temporal, and spatial variability of PLEs in >3,000 V. cholerae genomes collected between 1916 and 2019. (A) Timeline of PLE-positive strains colored by PLE variant type from 3,363 V. cholerae whole-genome sequences. The lower histogram provides the total number of strains per year, while the upper histogram represents a stacked percentage of those that are positive for each PLE. Because of the small number of sequenced genomes from isolates prior to 1960, those genomes were collapsed into a single column. Metadata for all strains analyzed can be found in Data Set S2. (B) Overview of the geographical occurrences of PLE(+) isolates by country of isolation. Each individual isolate is indicated with a colored dot corresponding to the PLE type, as in panel A, and grouped with all others for a given country. The relatively large numbers of isolates from Bangladesh are represented in the inset. (C) Global alignment of all 10 PLEs using progressiveMauve. The order was determined by phylogenetic distance between PLEs (Fig. S1). The mauve-colored regions are conserved across all PLE sequences, while the light gray regions are unique to that specific variant. All other regions vary in conservation between two or more PLEs and were assigned random colors from the variable palette. Annotated genes are shown to scale as black arrows, and several genes with known functions are labeled (§, lidI).
