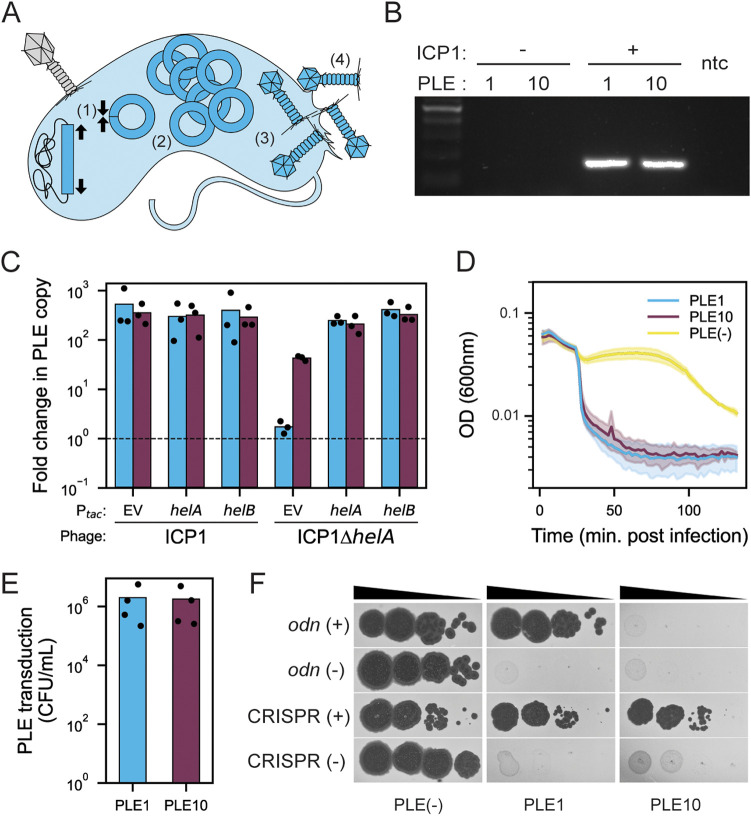
FIG 3.
PLE10 exhibits excision, replication, and packaging in response to ICP1 infection and evades the anti-PLE nuclease Odn. (A) Model showing the steps of the PLE-ICP1 interaction in a V. cholerae host cell. (1) The integrated PLE (in dark blue) excises and circularizes upon ICP1 infection. (2) PLE replicates and (3) is packaged into modified phage particles. (4) PLE transducing-particles are released through cell lysis, which occurs on an accelerated timeline. (B) Agarose gel of PCR products to detect circularized PLE in uninfected V. cholerae and following infection by ICP12006 lacking CRISPR-Cas (the phage isolate previously used to probe PLE circularization [19]). The lane on the far left is the ladder, and that on the far right is the no template control (ntc). (C) Quantification of change in PLE1 (blue) and PLE10 (purple) copy number 30 min after infection with ICP12006 lacking CRISPR-Cas or the ΔhelA mutant (the phage isolate previously used to probe PLE replication [18, 20, 29]). IPTG-inducible plasmid constructs (Ptac) were induced prior to phage infection. EV is the empty vector control. The dashed line indicates no change in copy number compared to the sample taken prior to phage infection (see Materials and Methods). (D) Lysis curves of V. cholerae harboring PLE1 or PLE10 following infection by ICP12006 lacking CRISPR-Cas (the phage isolate previously used to probe lysis kinetics [18, 22]) versus the PLE(−) control. (E) PLE-transducing particles generated during infection with ICP12006 lacking CRISPR-Cas (the phage isolate previously used to probe PLE transduction [18]). (F) Tenfold dilutions of the phage isolate carrying the anti-PLE locus indicated or mutant derivative spotted on V. cholerae with the PLE indicated (bacterial lawns are in gray, zones of killing are in black). Contemporary ICP1 isolates were used in this assay: the CRISPR(+) isolate with spacers against PLE1 and PLE10, ICP12017, was recovered from the same patient sample as the original PLE10(+) V. cholerae, and an Odn(+) isolate, ICP12019, were used. See Table S1 for a complete description of phage isolates. For experiments in panels B to F, PLEs were transduced into the same genetic background (V. cholerae E7946); see Table S1 for strain details.