FIG 4.
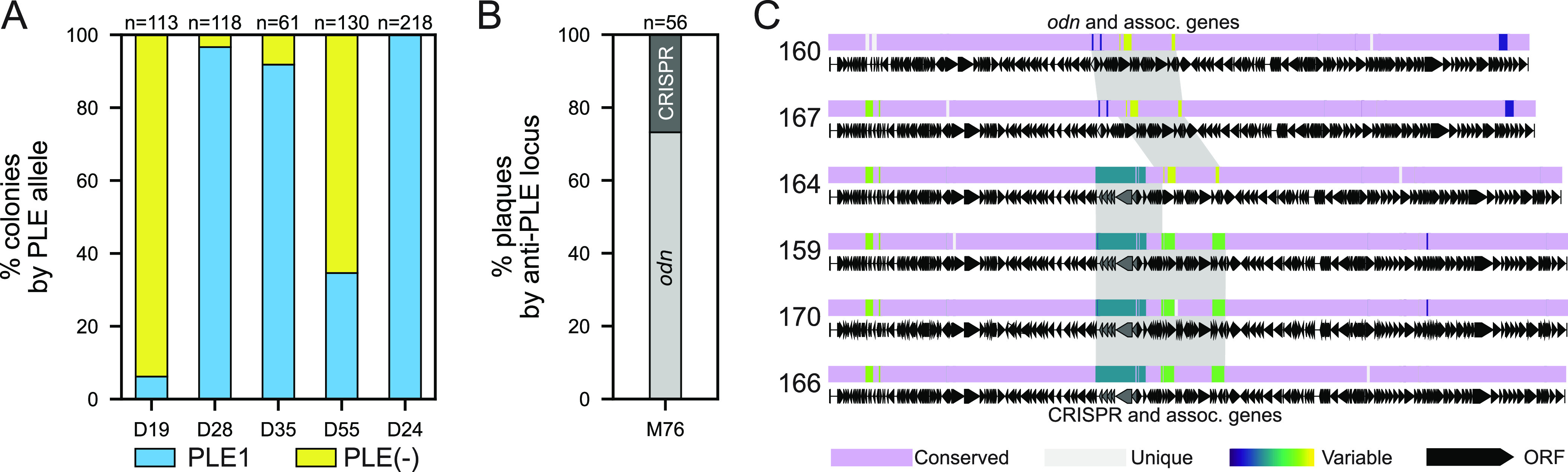
PLE(+)/PLE(−) V. cholerae and ICP1 anti-PLE loci heterogeneity within patients. (A) The fraction of total colonies with and without PLE for V. cholerae isolates within individual stool samples as determined by PCR. The number of colonies screened from each patient sample (given an arbitrary number by date of isolation with the prefix D are from icddr,b in Dhaka) are given above each bar. (B) The fraction of ICP1 plaques with the anti-PLE locus indicated recovered from one stool sample from Mathbaria (M76) as determined by PCR. (C) Alignment of six ICP1 isolates from stool sample M76 (panel B) using progressiveMauve. The mauve-colored regions are conserved across all ICP1 sequences, while the light gray regions are unique to that specific isolate. All other regions vary in conservation between two or more isolates and were assigned random colors from the variable palette. Annotated genes are shown as black arrows, and Cas and odn genes are shown in gray. The gray shading highlights regions with odn with associated genes (in phages 160 and 167) and CRISPR-Cas with associated genes (in phages 159, 170, and 166) at ≥95% sequence identity. Isolate 164 demonstrates recombination between the CRISPR-Cas region and the odn-associated region. See Table S1 for a complete description of phage isolates.
