FIG 3.
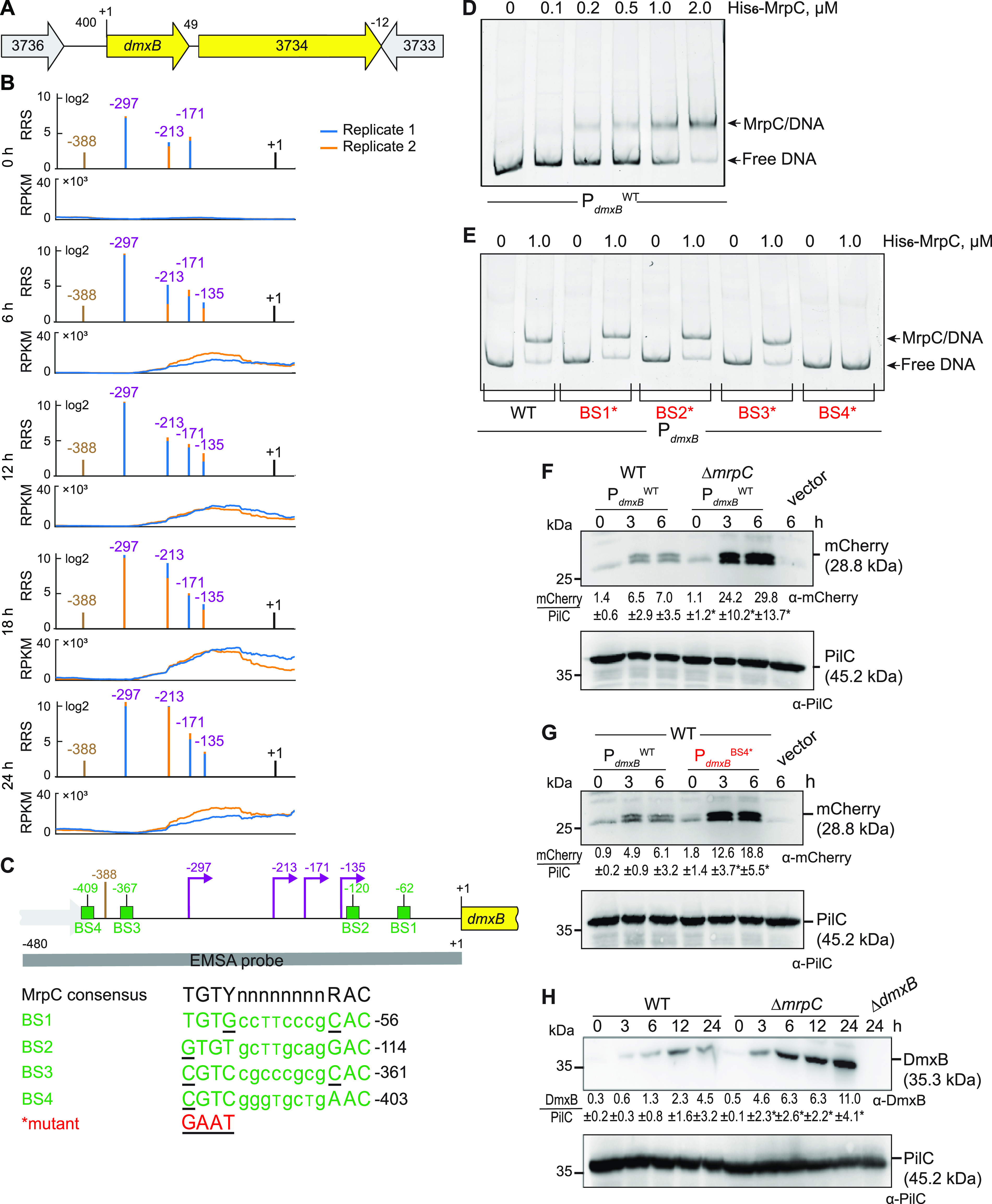
MrpC negatively regulates the expression of dmxB. (A) Schematic of the dmxB locus. The direction of transcription is indicated by the arrows. +1 indicates TSC of dmxB. Numbers above indicate the distance between start and stop codons of flanking genes. MXAN_3734 encodes a 577-amino acid residues protein with a C-terminal receiver domain of response regulators; the remainder of the protein does not contain known domains; MXAN_3734 is not important for development (40). (B) RNA-seq (bottom) and Cappable-seq (top) data at different time points. For each time point, data for both biological replicates are shown in blue and orange. For Cappable-seq, the RRS is indicated for each TSS on a log2 scale; for RNA-seq, reads per kilo base per million mapped reads (RPKM) values were calculated for each nucleotide position. Data from RNA-seq and Cappable-seq are from different samples. +1 indicates the dmxB TSC. TSSs as mapped by Cappable-seq are indicated in purple relative to the TSC of dmxB. The center of the MrpC ChIP-seq peak is indicated in brown. (C) Feature map of dmxB promoter region. +1 and color code is as in panel B. Green boxes labeled BS1 to 4 indicate potential MrpC binding sites based on the consensus sequence as defined by reference 50; sequences of BS1 to 4 are shown below and in which underlined text indicates a mismatch. Red indicates the sequence used to generate the mutant binding sites. The gray line indicates the EMSA probe and contains all four predicted MrpC binding sites. (D and E) MrpC binds to the dmxB promoter region using BS4. The indicated Hex-labeled probes were mixed with the indicated concentrations of His6-MrpC EMSA and analyzed by EMSA. (F) MrpC represses dmxB promoter(s). Total cell lysates from the indicated strains expressing mcherry from PdmxBWT were harvested from cells developed in MC7 submerged cultures at the indicated time points. A total of 10 μg of protein was loaded per lane, and samples were separated by SDS-PAGE. Top and bottom blots were probed with α-mCherry and α-PilC antibodies, respectively. The PilC blot served as a loading control. Numbers below the top panel indicate in the accumulation of mCherry relative to PilC as mean ± SD as measured in three biological replicates (Materials and Methods). *, P < 0.05; in Student’s t test in which samples from the ΔmrpC mutant were compared with samples from WT at the same time point. Vector with mCherry but without the dmxB promoter served as a negative control (vector). mCherry separates into two bands; the reason for this result is not known. (G) BS4 is important for MrpC-dependent repression of dmxB promoter(s). Total cell lysates from the indicated WT strains expressing mCherry from the two indicated promoters were prepared and analyzed as in panel F. (H) DmxB accumulates at increased levels in the ΔmrpC mutant. Total cell lysates of the indicated strains were harvested from cells developed in MC7 submerged conditions at indicated time points and analyzed as in panel F except that the accumulation of DmxB relative to PilC was calculated.
