FIG 6.
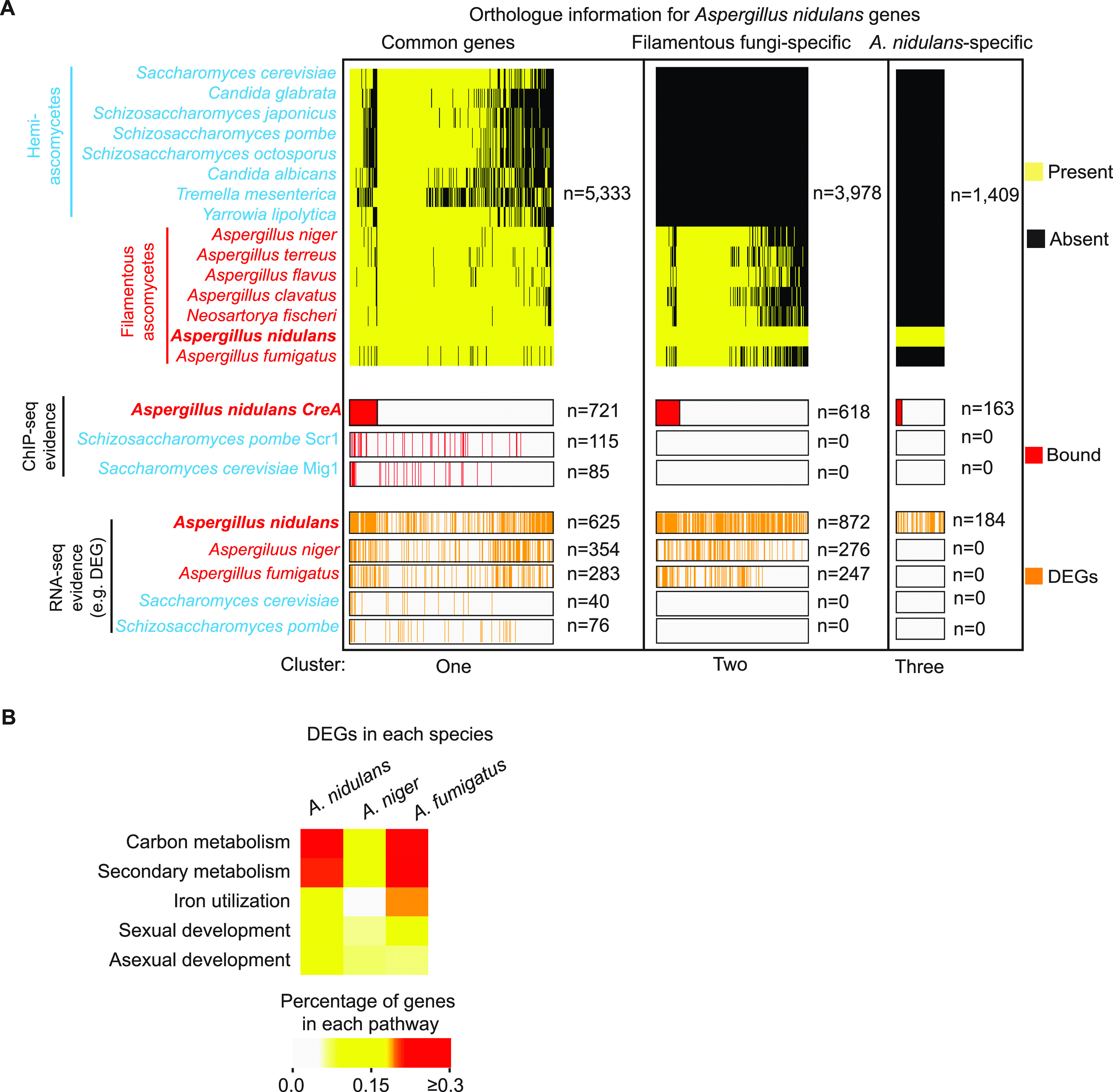
CreA targets many filamentous fungus-specific genes. (A) A summary diagram displaying orthologue information for A. nidulans genes in the indicated hemi-ascomycete and filamentous ascomycete species. The upper panel shows the presence and absence of orthologue for each A. nidulans gene in the indicated species. The A. nidulans genes were separated into three groups, namely, genes shared by most species (common), genes unique to filamentous ascomycetes (filamentous fungus specific), and genes unique to A. nidulans (A. nidulans specific). The middle panel marks the genes bound by CreA, S. pombe Scr1, or S. cerevisiae Mig1 based on ChIP evidence, while the lower panel indicates differentially expressed genes (as determined by RNA-seq) in the mutant of the creA homologue of the corresponding species. Gene number for each category is indicated by n. (B) Heatmap plot showing the percentage of DEGs enriched in the indicated pathways for the creAΔ mutant of the three different species.
