Abstract
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has infected more than 180 million people since the onset of the pandemic. Despite similar viral load and infectivity rates between children and adults, children rarely develop severe illness. Differences in the host response to the virus at the primary infection site are among the mechanisms proposed to account for this disparity. Our objective was to investigate the host response to SARS-CoV-2 in the nasal mucosa in children and adults and compare it with the host response to respiratory syncytial virus (RSV) and influenza virus. We analyzed clinical outcomes and gene expression in the nasal mucosa of 36 children with SARS-CoV-2, 24 children with RSV, 9 children with influenza virus, 16 adults with SARS-CoV-2, and 7 healthy pediatric and 13 healthy adult controls. In both children and adults, infection with SARS-CoV-2 led to an IFN response in the nasal mucosa. The magnitude of the IFN response correlated with the abundance of viral reads, not the severity of illness, and was comparable between children and adults infected with SARS-CoV-2 and children with severe RSV infection. Expression of ACE2 and TMPRSS2 did not correlate with age or presence of viral infection. SARS-CoV-2–infected adults had increased expression of genes involved in neutrophil activation and T-cell receptor signaling pathways compared with SARS-CoV-2–infected children, despite similar severity of illness and viral reads. Age-related differences in the immune response to SARS-CoV-2 may place adults at increased risk of developing severe illness.
Keywords: COVID-19, pneumonia, viral infections, IFNs, T cells
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is responsible for the current pandemic of coronavirus disease (COVID-19). A notable feature of SARS CoV-2 infection is the dramatic heterogeneity in clinical phenotypes. Although most infected individuals remain asymptomatic or develop mild disease, some develop severe SARS-CoV-2 pneumonia and acute respiratory distress syndrome (ARDS). These severe infections account for most of the public health and societal impacts of the COVID-19 pandemic and disproportionately affect the elderly while mostly sparing children (1–3). In contrast, other common viral respiratory infections, including respiratory syncytial virus (RSV) and influenza virus (IV), tend to be most severe in young children and elderly adults. Of the approximately 225,000 deaths from COVID-19 reported in the first 8 months of the pandemic in the United States, only 189 occurred in children less than 18 years of age (4), despite at least 277,285 documented infections in children over the same period (5). Identifying the mechanisms driving morbidity and mortality differences between children and adults infected with SARS-CoV-2 may provide essential insights for treating patients with COVID-19.
SARS-CoV-2 infection begins in the nasal mucosa (6, 7), where the spike protein binds angiotensin-converting enzyme 2 (ACE2) on the surface of epithelial cells. In concert with host proteases, principally transmembrane serine protease 2 (TMPRSS2), the virus enters the cell to begin replication (8–10). Initial studies suggested that ACE2 expression in the airways increases with age (11, 12), which could lead to lower infectivity and lower viral load in children. However, subsequent studies have demonstrated that SARS-CoV-2 viral load is similar in children and adults, suggesting the relative protection from COVID-19 in children is not due to age-related differences in infectivity (13–16). As there are important differences between the pediatric and adult immune systems, the host response to SARS-CoV-2 could explain the observed differences in clinical phenotypes in children and adults. The primary host response to viral infection is antiviral IFN signaling to limit viral replication, recruit immune cells, and clear infected cells (17). However, some viruses have developed strategies to evade this response, and SARS-CoV-2 is known to inhibit host IFN responses in vitro (18, 19). This observation has led investigators to speculate that differences in antiviral IFN signaling at the primary infection site might be responsible for differences in the frequency of severe COVID-19 between children and adults. To date, no studies have compared the host response to the virus at the primary site of SARS-CoV-2 infection in mildly symptomatic children and adults.
This study aimed to assess the host response to SARS-CoV-2 at the primary site of infection in children and adults and how it may differ from the host response to other respiratory viruses, namely RSV and IV, that cause severe disease in children. Curettage and transcriptional profiling of the nasal mucosa can provide valuable insights into host–pathogen interactions at the initial infection site that may drive disease outcomes (20–23). Therefore, we performed RNA sequencing (RNA-seq) of nasal mucosa samples from children and adults with SARS-CoV-2, children with RSV or IV, and healthy children and adults. Local antiviral IFN signaling did not differ between children and adults infected with SARS-CoV-2 and was similar to children with severe RSV infection. Viral load, but not severity of illness, correlated with the magnitude of the IFN response. Notably, expression of genes involved in neutrophil activation and T-cell receptor signaling was increased in adults compared with children with asymptomatic or mild SARS-CoV-2 infection. Our findings suggest that the local IFN response does not drive differences in the clinical phenotypes associated with SARS-CoV-2 infection. Rather, age-related differences in the innate and adaptive immune response to SARS-CoV-2 infection in the upper respiratory tract may be critical risk factors for severe disease.
Some of the results of these studies have been previously reported in the form of a preprint (https://doi.org/10.1101/2021.01.26.21250269).
Methods
Study Population
Our study included the following six groups: healthy children, children infected with SARS-CoV-2, children infected with RSV, children infected with IV, healthy adults, and adults infected with SARS-CoV-2. Approval for this study was obtained from the institutional review board at Ann and Robert H. Lurie Children’s Hospital of Chicago. Informed consent was obtained from adult participants and guardians of pediatric participants. The medical records of all pediatric patients admitted between December 2019 and November 2020 were screened for inclusion criteria and the absence of exclusion criteria (see below). We sequenced samples from healthy children recruited for a previous study as negative controls. These samples were obtained from healthy children attending a well-child check at Ann and Robert H. Lurie Children’s Hospital of Chicago Outpatient Center in May 2018. If the patient was eligible for the study, a study team member approached for consent. For SARS-CoV-2–infected children, both the child and accompanying adult were invited to participate.
Inclusion Criteria
Children were eligible for inclusion in the SARS-CoV-2–infected group if they had a positive PCR test for SARS-CoV-2 at admission to the hospital or our drive-through testing center (Figure E1A in the data supplement). Symptoms were collected based on reports by participants or their guardians.
Adults accompanying a SARS-CoV-2–infected child or who were themselves admitted and cared for in our pediatric hospital with a positive PCR test for SARS-CoV-2 were eligible for inclusion (Figure E1B). PCR testing was not available for the adults accompanying a child before enrollment. We categorized adult participants as infected with SARS-CoV-2 if RNA-seq detected SARS-CoV-2 RNA. Adults without detectable SARS-CoV-2 via RNA-seq were classified as healthy controls. Self-reported symptoms were collected from adults at the time of enrollment and via follow-up weekly phone calls for 3 weeks. Adults were asked about 11 of the most common symptoms reported with SARS-CoV-2 infection (24).
Participants were eligible for inclusion in the viral positive control group if they had a PCR confirming infection with RSV or IV, were admitted to the pediatric intensive care unit (PICU), and had symptoms of pulmonary parenchymal disease, including any of the following: 1) infiltrates, areas of atelectasis, or hyperinflation on chest radiograph; 2) rales, wheezing, retractions, tachypnea, or any form of respiratory distress on clinical exam; or 3) an oxygen requirement to maintain peripheral oxygen saturation levels above 95%.
Healthy pediatric controls were eligible for inclusion if they were less than 18 years of age and had no signs or symptoms of viral infection at the time of their scheduled well-child visit with their primary care provider (Figure E1C). All healthy pediatric controls were clinically stable without apparent viral or bacterial infection when the nasal epithelial scrapings were obtained. In addition, metagenomic analysis of RNA-seq data using the OneCodex platform did not detect any viral reads for 29 common human respiratory viruses in any of the healthy control samples. Healthy controls were also followed for 2 weeks via electronic medical record to ensure no infections occurred within that time frame.
Exclusion Criteria
Subjects were not eligible for inclusion in the pediatric healthy control group if they had any significant medical comorbidities, including preexisting lung disorders, preexisting cardiac disease, immunodeficiency, malignancy, and neurologic disorders increasing the risk of aspiration or respiratory failure. Children with a clinical concern for multisystem inflammatory syndrome in children (MIS-C) were not eligible for inclusion in the SARS-CoV-2 cohort.
Patient Enrollment
We enrolled 170 participants in the study (Figure E1). Before sample collection, 10 participants withdrew consent. Eight participants were discharged before sample collection. After the isolation of RNA, 26 samples were excluded owing to low RNA quality. Following RNA-seq, an additional 14 samples were excluded: 1 for low-quality RNA-seq library, 3 for sex mismatch between biological sex indicated in the research records and expression of XIST or RPS4Y1 genes, 3 for negative PCR testing results, 1 for MIS-C diagnosis, 1 for unavailable patient research records, and 5 healthy controls for detection of viral reads for common respiratory viruses. A cohort of 105 participants (7 healthy children, 36 children infected with SARS-CoV-2, 24 children infected with RSV, 9 children infected with IV, 13 healthy adults, and 16 adults infected with SARS-CoV-2) was used for the analysis.
Clinical Data
Clinical data were collected and managed using REDCap electronic data capture tools hosted at the Northwestern University Clinical and Translational Sciences Institute (25). Hospital length of stay was determined from admission and discharge dates. Readmissions within 24 hours of discharge were considered a continuation of hospitalization. If a patient had multiple PICU stays during a single encounter, the PICU length of stay was cumulative. Advanced respiratory support was defined by the use of high-flow nasal cannula (HFNC), noninvasive positive pressure ventilation (NIPPV), or invasive mechanical ventilation (IMV). Respiratory support and oxygen duration were determined by documentation on a calendar day of nonhome advanced respiratory support or oxygen to exclude chronic baseline support. Pediatric ARDS was defined using the Pediatric Acute Lung Injury Consensus Conference criteria (26). Detection of a copathogen was defined as a positive viral PCR or any positive bacterial or fungal cultures.
Nasal Curettage and RNA Isolation
Owing to infection control procedures during the COVID-19 pandemic, nasal mucosa curettage was performed during routine care by healthcare providers caring for the patient. Providers received training just before sample collection by trained study personnel. Nasal epithelial cells were obtained by mucosal scrape biopsy of the inferior turbinate using a sterile plastic curette (Rhino-Pro curette; Arlington Scientific) as previously described (27) to obtain a predominantly epithelial cell population. A total of three single-pass scrapings per nares were performed per subject. The curette tips were cut and placed into RNase-free collection tubes containing 350 μL of RLT buffer (Qiagen) supplemented with 2-mercaptoethanol, vortexed vigorously, and stored at −80°C. RNA was isolated using Qiagen RNeasy extraction kits.
RNA Sequencing and Analysis
RNA quality and quantity were assessed using 4200 TapeStation High Sensitivity RNA tapes (Agilent), and RNA-seq libraries were prepared from 1 ng of total RNA using SMARTer Stranded Total RNA-seq Kit v2 (Takara Bio). After quality control using 4200 TapeStation High Sensitivity DNA tapes (Agilent), dual-indexed libraries were pooled and sequenced on a NextSeq 500 instrument (Illumina), 75 cycles, single-end, to an average sequencing depth of 5.88M reads.
FASTQ files were generated using bcl2fastq (Illumina). Viral RNA was detected using a custom hybrid genome prepared by joining FASTA, GFF, and GTF files for GRCh37.87, SARS-CoV-2 (NC_045512.2), Influenza A/California/07/2009 (GCF_001343785.1), and RSV/S2 ts1C (GCF_000856445.1) genomes. An additional negative-strand transcript spanning the entirety of the SARS-CoV-2 genome was then added to the GTF and GFF files to enable detection of SARS-CoV-2 replication. To facilitate reproducible analysis, samples were processed using the publicly available nf-core/RNA-seq pipeline version 1.4.2 implemented in Nextflow 19.10.0 using Singularity 3.2.1–1 with the minimal command nextflow run nf-core/rnaseq -r 1.4.2 –singleEnd -profile singularity –reverseStranded –three_prime_clip_r2 3 (28–30). Briefly, lane-level reads were trimmed using trimGalore! 0.6.4 and aligned to the hybrid genome described above using STAR 2.6.1 d (31). Gene-level assignment was then performed using featureCounts 1.6.4 (32). First, putative sample swaps were identified by comparing known patient sex with sex determined by XIST and RPS4Y1 expression levels. No mismatched samples were included in the analysis. For pairwise comparison, EdgeR was used to identify differentially expressed genes between children and adults with mild disease (33, 34). Gene ontology (GO) analysis was run using GOrilla (35).
Weighted Gene Coexpression Network Analysis
Weighted gene coexpression network analysis (WGCNA) was performed using WGCNA version 1.69 with default settings unless otherwise noted (36). Highly variable genes among all participant samples (n = 105) and pediatric participant samples only (n = 76) were included in analyses. To best capture patterns of coregulation, a signed network was used. Using the pickSoftThreshold function, we empirically determined a soft threshold of 3 to best fit the network structure. A minimum module size of 30 genes was chosen. Module eigengenes were then related to patient and sample metadata using biweight midcorrelation. Module GO enrichment was determined using GOrilla (35). Gene set enrichment analysis (GSEA) was run using weighted, preranked setting based on log2 fold change values reported by edgeR (37).
Deconvolution of Bulk RNA-Seq Signatures
Deconvolution of bulk RNA-seq signatures was performed using AutoGeneS version 1.0.3 (38). For details of the functions used and their parameters, see the code available at https://github.com/NUPulmonary/2021_Koch. Briefly, we used a single-cell RNA-seq dataset from Ordovas-Montanes and colleagues (39), which contains nasal epithelial and immune cells, to train the AutoGeneS model. AutoGeneS selects cell type-specific genes by both minimizing the correlation and maximizing the distance between the cell type-specific average gene expression profiles. The model was then applied to bulk RNA-seq data to estimate specific cell types’ proportions using regression.
Statistical Analysis
Categorical data were summarized by percentages and compared using Fisher’s exact test. Continuous, nonparametric data were summarized using medians with interquartile ranges and compared using Mann-Whitney U or Kruskal-Wallis testing. The Benjamini-Hochberg procedure was used for false discovery rate correction when performing multiple comparisons. Data were considered statistically significant at P-adjusted < 0.05. Log-transformed counts per million (CPM) data were shown in graphs for visualization purposes. Statistical analyses were performed using GraphPad Prism version 9.0.0 and R version 4.0.3. Data were visualized using ggplot2 version 3.3.1 and GraphPad Prism.
Data and Code Availability
Processed RNA-seq counts are available as Table E1. Raw data is in the process of being deposited to dbGaP/SRA. Code is available via https://github.com/NUPulmonary/2021_Koch.
Results
Description of the Cohort
The demographics of the pediatric and adult cohorts are summarized in Table 1 and Table E1. When considering only children, patients with RSV were younger, and a larger proportion was previously healthy than those with SARS-CoV-2 (Table 1). The ages and proportion of individuals with chronic medical conditions in the uninfected adult group were no different from those in the SARS-CoV-2–infected adult group (Table E1).
Table 1.
Description of Pediatric Cohort
| SARS-CoV-2 Children |
RSV Children |
IV Children |
Adjusted P Value | |
|---|---|---|---|---|
| n | 36 | 24 | 9 | |
| Median age, yr (IQR) | 1.9 (0.4–15.0) | 0.33 (0.16–0.44) | 1.7 (1.4–7.0) | 0.003 |
| Sex | ||||
| Male, n (%) | 18 (50.0) | 10 (41.7) | 5 (55.6) | 0.92 |
| Female, n (%) | 18 (50.0) | 14 (58.3) | 4 (44.4) | |
| Race | ||||
| White, n (%) | 8 (22.2) | 13 (54.2) | 1 (11.1) | 0.03 |
| African American, n (%) | 4 (11.1) | 1 (4.2) | 2 (22.2) | |
| Asian, n (%) | 0 (0.0) | 2 (8.3) | 1 (11.1) | |
| Other, n (%) | 23 (63.9) | 6 (25.0) | 4 (44.4) | |
| Unspecified, n (%) | 1 (2.8) | 2 (8.3) | 1 (11.1) | |
| Ethnicity | ||||
| Non-Hispanic, n (%) | 11 (30.6) | 15 (62.5) | 4 (44.4) | 0.14 |
| Hispanic, n (%) | 24 (66.7) | 7 (29.2) | 4 (44.4) | |
| Unspecified, n (%) | 1 (2.8) | 2 (8.3) | 1 (11.1) | |
| Medical history | ||||
| Healthy, n (%) | 12 (33.3) | 19 (79.2) | 3 (33.3) | 0.006 |
| Respiratory, n (%) | 9 (25.0) | 2 (8.3) | 5 (55.6) | 0.06 |
| Cardiovascular, n (%) | 5 (13.9) | 1 (4.2) | 1 (11.1) | 0.82 |
| Neurologic, n (%) | 6 (16.7) | 1 (4.2) | 2 (22.2) | 0.47 |
| Hematologic, n (%) | 0 (0.0) | 1 (4.2) | 0 (0) | 0.76 |
| Oncologic, n (%) | 2 (5.6) | 0 (0) | 0 (0) | 0.88 |
| Chronic kidney disease, n (%) | 1 (2.8) | 1 (4.2) | 0 (0) | 1 |
| Gastrointestinal, n (%) | 8 (22.2) | 2 (8.3) | 2 (22.2) | 0.62 |
| Endocrine, n (%) | 3 (8.3) | 2 (8.3) | 1 (11.1) | 1 |
| Metabolic/genetic, n (%) | 8 (22.2) | 1 (4.2) | 2 (22.2) | 0.38 |
| Presentation* | ||||
| Respiratory, n (%) | 20 (57.1) | 24 (100) | 9 (100) | — |
| Gastrointestinal, n (%) | 3 (8.6) | 0 (0) | 0 (0) | — |
| Fever only, n (%) | 5 (14.3) | 0 (0) | 0 (0) | — |
| Nonrespiratory viral syndrome, n (%) | 2 (5.7) | 0 (0) | 0 (0) | — |
| Asymptomatic, n (%) | 5 (14.3) | 0 (0) | 0 (0) | — |
| Median day of illness at collection (IQR) | 4.0 (2.0–6.8) | 4.0 (3.0–4.3) | 5.0 (4.0–5.0) | 0.88 |
| Severity | ||||
| ICU admission, d (%) | 8 (22.2) | 24 (100) | 9 (100) | — |
| Median ICU LoS, d (IQR) | 3.0 (2.0–6.0) | 7.0 (4.0–10.0) | 6.0 (3.0–9.0) | 0.68 |
| Median Hospital LoS, d (IQR) | 3.0 (2.0–6.0) | 7.0 (5.0–12.0) | 6.0 (3.0–9.0) | 0.006 |
| ARDS, n (%) | 1 (2.8) | 3 (12.5) | 1 (11.1) | 0.62 |
| Vasoactive medications, n (%) | 1 (2.8) | 2 (8.3) | 0 (0) | 0.89 |
| Respiratory support | ||||
| Collection | ||||
| None, n (%) | 29 (80.6) | 1 (4.2) | 0 (0) | 3.7 × 10–13 |
| Low flow interface, n (%) | 4 (11.1) | 0 (0) | 0 (0) | |
| HFNC, n (%) | 1 (2.8) | 13 (54.2) | 3 (33.3) | |
| NIPPV, n (%) | 2 (5.6) | 1 (4.2) | 4 (44.4) | |
| IMV, n (%) | 0 (0.0) | 9 (37.5) | 2 (22.2) | |
| Oxygen, n (%) | 7 (19.4) | 23 (95.8) | 9 (100) | 1 |
| Peak | ||||
| None, n (%) | 26 (72.2) | 0 (0) | 0 (0) | 1.4 × 10–11 |
| Low flow interface, n (%) | 3 (8.3) | 0 (0) | 0 (0) | |
| HFNC, n (%) | 3 (8.3) | 12 (50.0) | 3 (33.3) | |
| NIPPV, n (%) | 4 (11.1) | 2 (8.3) | 4 (44.4) | |
| IMV, n (%) | 0 (0.0) | 10 (41.7) | 2 (22.2) | |
| Oxygen, n (%) | 9 (25.0) | 24 (100) | 9 (100) | 1 |
| Median respiratory support days† (IQR) | 5.0 (2.0–6.0) | 6.5 (4.0–11.0) | 5.0 (3.0–8.0) | 0.62 |
| Median supplemental oxygen days (IQR) | 3.0 (2.0–6.0) | 7.0 (4.8–10.0) | 5.0 (3.0–9.0) | 0.26 |
| Copathogen detection | ||||
| Virus, n (%) | 0 (0.0) | 3 (12.5) | 1 (11.1) | 0.17 |
| Bacteria, n (%) | 0 (0.0) | 6 (25.0) | 1 (11.1) | 0.02 |
Definition of abbreviations: ARDS = acute respiratory distress syndrome; HFNC = high-flow nasal canula; IMV = invasive mechanical ventilation; IQR = interquartile range, IV = influenza virus; LoS = length of stay, NIPPV = noninvasive positive pressure ventilation; RSV = respiratory syncytial virus; SARS-CoV-2 = severe acute respiratory syndrome coronavirus 2.
Adjusted P values were calculated by Kruskal-Wallis test for medians and Fisher’s exact test for proportions with Benjamini-Hocherg false discovery rate correction. Values less than 0.05 were considered statistically significant.
n = 35 for SARS-CoV-2 presentation owing to incomplete medical records.
Cumulative days requiring HFNC, NIPPV, or IMV.
Figure 1A summarizes the clinical course of the hospitalized pediatric participants infected with IV, RSV, or SARS-CoV-2. There was no difference in the time from symptom onset to nasal sample collection between the groups (Figure 1B). Consistent with their inclusion as positive controls for moderate and severe viral respiratory tract infections, children with RSV were hospitalized longer and received higher respiratory support levels than children with SARS-CoV-2 (Figures 1C and 1D and Table 1). Similarly, children with IV received increased respiratory support than children with SARS-CoV-2 (Figure 1D and Table 1). Of the 36 children with SARS-CoV-2, 35 were admitted to the hospital and 8 required ICU-level care. Of those eight participants, seven were admitted with respiratory symptoms requiring noninvasive respiratory support, whereas one was admitted with severe diabetic ketoacidosis in the setting of fever. None of the SARS-CoV-2–positive children were intubated. However, one patient with a history of obesity (body mass index 37.2) met the pediatric ARDS criteria while supported on full-face bilevel positive airway pressure. The age, sex, race, and ethnicity of the children with IV and the uninfected children did not differ from those with SARS-CoV-2 (Figures E1D and E1F–E1I). A larger proportion of children with SARS-CoV-2 identified as Hispanic compared with RSV (Figure E1H). The sex, race, and ethnicity were not different between uninfected and infected adult groups (Figures E1E-E1I and Table E1).
Figure 1.
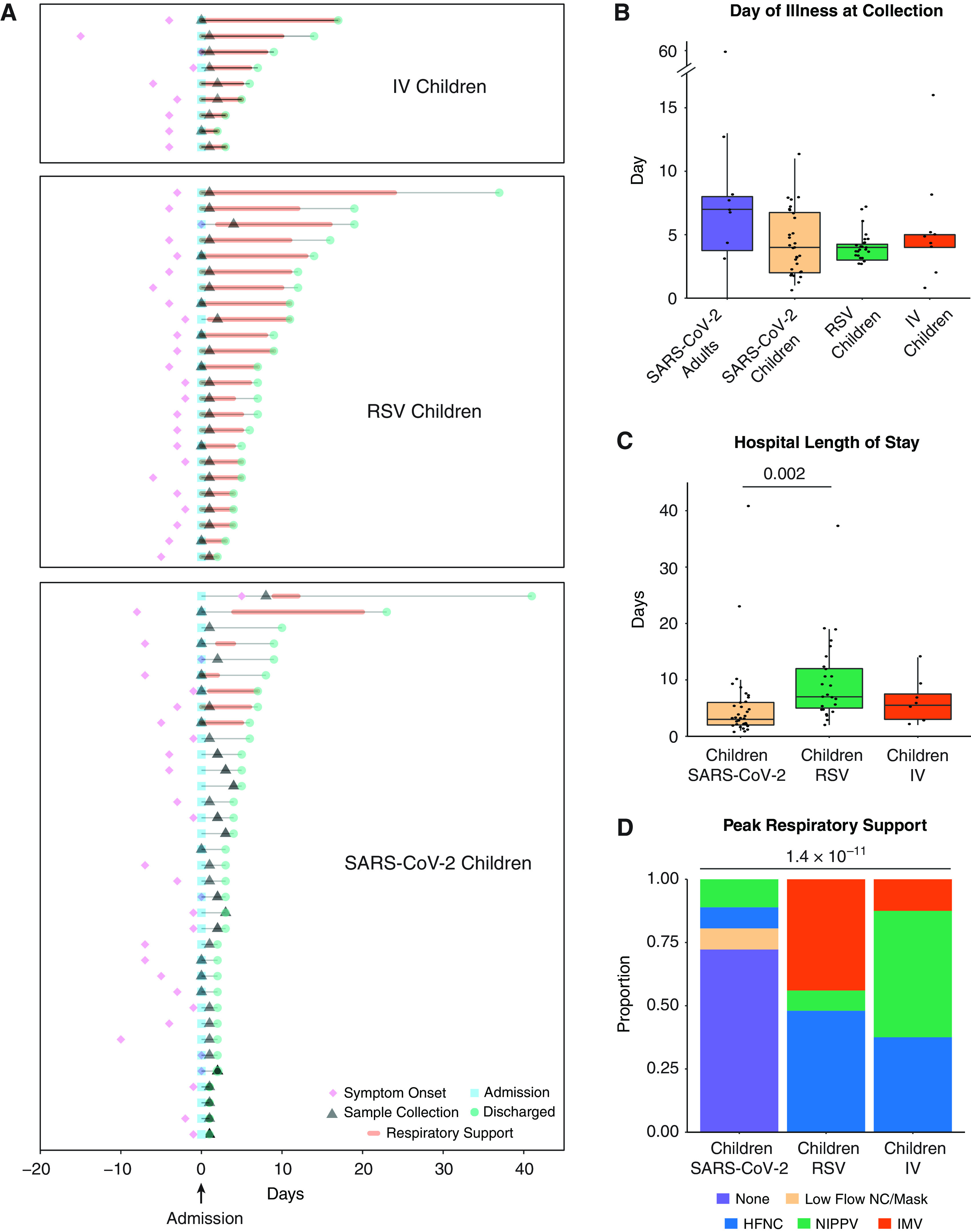
Description of the cohort. (A) Clinical course of pediatric participants. Timing of symptom onset (diamond), admission (square), sample collection (triangle), discharge (circle), hospital stay (thin line), and duration of advanced (HFNC, NIPPV, or IMV) respiratory support (bold line) of hospitalized children with severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) (n = 36), RSV (n = 24), and IV (n = 9). (B) Self- or parent/guardian-reported time since symptom onset at the time of sample collection was similar between all participant groups. Pairwise comparisons of medians performed using the Mann-Whitney U test. (C) Children with RSV infection had a longer hospital length of stay than children with SARS-CoV-2 infection. Pairwise comparisons of medians performed using the Mann-Whitney U test. (D) Higher proportions of children with IV and RSV infection required HFNC, NIPPV, and IMV at peak illness severity when compared with children with SARS-CoV-2 infection. Proportions compared using Fisher’s exact test. All P values were adjusted using Benjamini-Hochberg FDR correction. Differences were not significant (P-adjusted > 0.05) unless noted. FDR = false discovery rate; HFNC = high flow nasal cannula; IMV = intermittent mechanical ventilation; IV = influenza virus; NIPPV = noninvasive positive pressure ventilation; RSV = respiratory syncytial virus.
The majority of the adult participants with SARS-CoV-2 reported mild illness (Table E2). There was no difference in the time from symptom onset to nasal sample collection between adults and all pediatric groups (Figure 1B). Two adult participants (ages 19 and 20 yr) were admitted to our pediatric hospital at the time of enrollment. An additional adult participant reported admission to an outside hospital after enrollment in our study. The two patients admitted to our hospital had respiratory illnesses requiring admission to the PICU for noninvasive positive pressure ventilation with supplemental oxygen. Neither met the ARDS criteria. The participant admitted to an outside hospital reported an admission duration of 3 days and did not require ICU-level care.
The symptoms reported by participants or the guardian for children with SARS-CoV-2 infection are summarized in Table E1. Of the 16 adults with SARS-CoV-2 infection (see Methods), 7 (44%) were asymptomatic, and the other 9 (56%) reported symptoms. The majority of children with SARS-CoV-2 infection experienced respiratory symptoms (57%). Other symptoms included fever only (14%), gastrointestinal symptoms (9%), nonrespiratory viral syndromes (6%), and asymptomatic (14%) (Table 1). The asymptomatic children with SARS-CoV-2 were detected on routine screening at admission.
SARS-CoV-2 Viral Load and Expression of SARS-CoV-2 Entry Factors in the Nasal Mucosa Do Not Correlate with Age
To characterize our participant sample data and confirm clinical diagnoses, we queried bulk RNA-seq data obtained from curettage of nasal mucosa for the presence of SARS-CoV-2, RSV, or IV reads. Viral reads for SARS-CoV-2 were detected in 28 of 36 samples collected from children diagnosed with SARS-CoV-2 infection by PCR performed in the hospital diagnostic laboratory (Figure 2A). Of the 24 children diagnosed with RSV, all had detectable RSV reads in RNA-seq data. Six out of 9 children with IV had detectable influenza A viral reads in their nasal mucosa. Of the 29 adults accompanying children with SARS-CoV-2 infection, 14 had detectable SARS-CoV-2 reads, and 2 young adults admitted to the pediatric hospital with a positive SARS-CoV-2 PCR test did not have detectable viral reads. Nine samples were obtained from healthy children. RNA-seq did not detect any SARS-CoV-2, RSV, or IV reads in these samples; however, a more in-depth analysis of these samples using the TWIST viral panel as implemented in the OneCodex platform demonstrated the presence of enterovirus and parainfluenza virus in two of the samples. These samples were excluded from subsequent analysis, leaving seven subjects in the healthy control group (Figure E1C).
Figure 2.
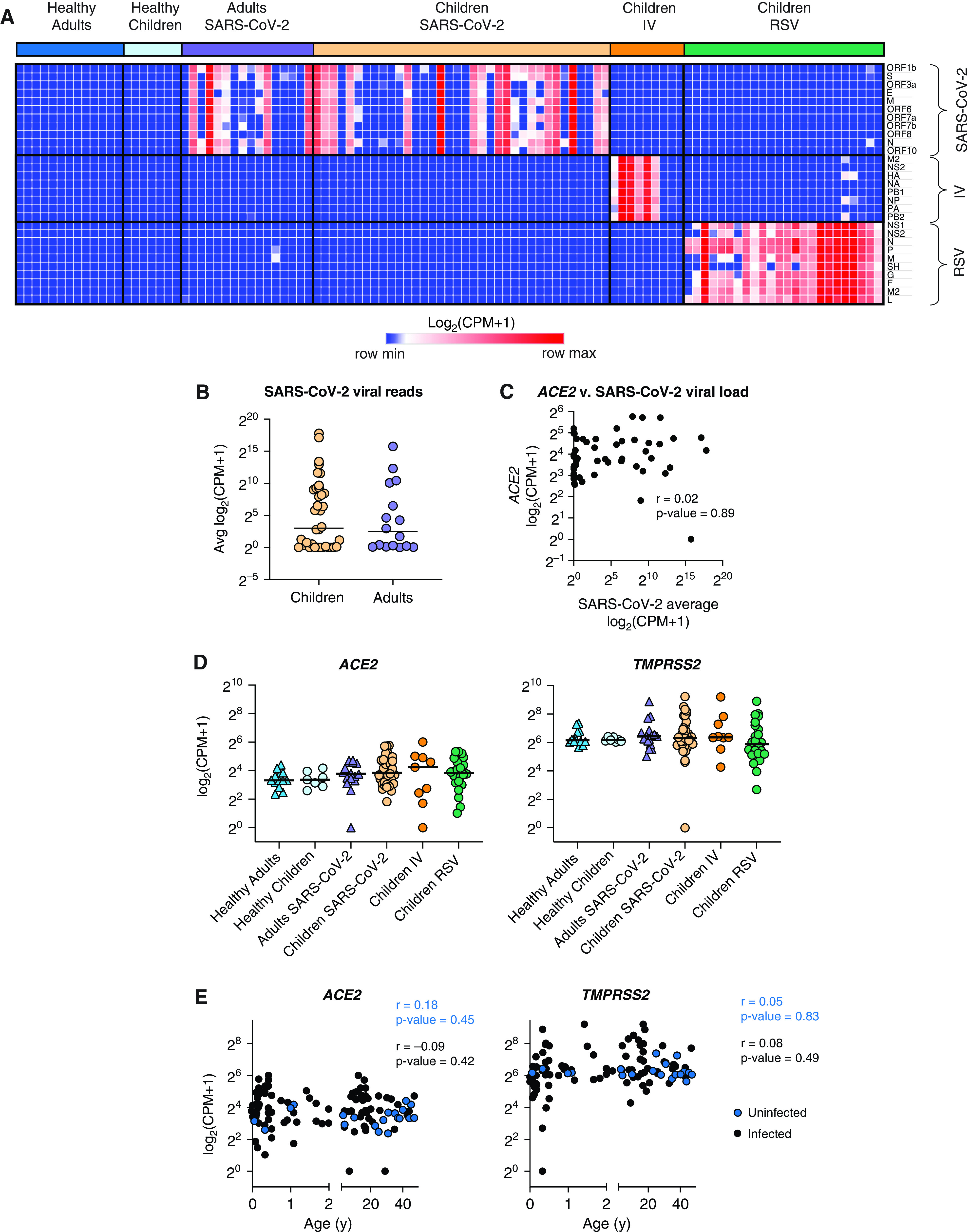
SARS-CoV-2 viral load and expression of SARS-CoV-2 entry factors in the nasal mucosa does not correlate with age. We compared the nasal transcriptome of 36 children with SARS-CoV-2 infection with 7 healthy children, 24 children with RSV infection, 9 children with IV infection, 16 adults with SARS-CoV-2 infection, and 13 healthy adults. (A) Detection of viral reads for SARS-CoV-2, influenza A/California/07/2009, and RSV/S2 ts1C in RNA-sequencing data for all participant samples (n = 105). Heatmap of viral genes for SARS-CoV-2, IV, and RSV. Log-transformed pseudocounts (counts per million [CPM] + 1) are shown. (B) Normalized reads were averaged for all 11 SARS-CoV-2 viral genes. Average SARS-CoV-2 reads did not differ between children (yellow circle) and adults (purple triangle). Log-transformed pseudocounts (CPM + 1) are shown. (C) Averaged SARS-CoV-2 viral reads did not correlate with ACE2 expression. Log-transformed pseudocounts (CPM + 1) are shown. Pearson correlation coefficient (r) and P value are shown. (D) Normalized expression of ACE2 and TMPRSS2 was similar across all participant groups; differences not significant, Kruskal-Wallis testing with Benjamini-Hochberg FDR correction; all adjusted P values > 0.05. Log-transformed pseudocounts (CPM + 1) are shown. (E) ACE2 and TMPRSS2 expression did not correlate with age in uninfected nor infected participants. Pearson correlation coefficients (r) and P values are shown. Log-transformed pseudocounts (CPM + 1) are shown.
In agreement with previous reports (13–16), we found that SARS-CoV-2 viral load was similar between children and adults and did not correlate with ACE2 expression (Figures 2B and 2C). Several studies have reported that the expression of ACE2 and TMPRSS2, two key entry factors for SARS-CoV-2, increases with age (11, 12). Although some reports suggested that IFNs produced in response to viral infection can lead to increased expression of ACE2, potentially contributing to the spread of infection (7, 40), other reports have found no correlation or the opposite effect (41–43). We found that the expression of ACE2 and TMPRSS2 was not different between healthy children and adults. Moreover, none of the viral infections led to increased expression of ACE2 and TMPRSS2 (Figure 2D). Similarly, ACE2 and TMPRSS2 expression did not correlate with age (Figure 2E). Taken together, transcriptomic data confirmed clinical diagnoses for SARS-CoV-2, IV, and RSV infections. Moreover, in contrast to previous studies, we did not find a correlation between SARS-CoV-2 entry factors and age in our cohort.
Cell-Type Deconvolution of Transcriptomic Signatures Identifies an Increase in Immune Cells within the Nasal Mucosa Associated with Viral Infection
Bulk RNA-seq data obtained from complex tissue samples composed of multiple cell types can obscure disease-specific signatures as RNA molecules from different cell types are averaged. Accordingly, our initial exploratory analysis did not reveal a distinct structure or pattern within our dataset (Figures E2A and E2B). To address the potential heterogeneity in cell-type composition in our samples, we performed in silico cell-type deconvolution of our bulk RNA-seq data using cell type-specific reference data from the nasal mucosa (39). Hierarchical clustering of predicted cell-type abundance distinguished six clusters of samples (Figure 3A and Table E2). Four clusters were characterized by an abundance of immune cells (clusters 1–4), whereas two clusters were enriched with secretory and ciliated epithelial cells (clusters 5 and 6). In line with previous reports (44, 45), deconvolution analysis demonstrated that most nasal mucosa samples from healthy participants were enriched for epithelial cells, with all of the samples from healthy children dominated by a ciliated cell signature (Figures 3A and 3B). In contrast, samples from children with RSV infection were equally divided between the secretory cell cluster and the immune cell clusters, consistent with previous reports of ciliated cells being replaced with secretory cells and the recruitment of neutrophils in RSV infection (46, 47). Increased immune cell abundance was observed in most samples from children infected with IV and SARS-CoV-2 (Figures 3A and 3B and Figure E2C). The time from onset of symptoms to sample collection did not differ between epithelial and immune dominant subsets (Figure 3C). In children and adults infected with SARS-CoV-2, viral reads were increased in immune-enriched samples compared with epithelial-enriched samples, suggesting increased viral load is associated with immune cell recruitment to the nasal mucosa (Figure 3D). To confirm that differences in cell-type composition were not obscuring differences in ACE2 expression in children and adults, we analyzed ACE2 expression in samples from the epithelial cell clusters and the immune cell clusters separately. There remained no difference in ACE2 expression between children and adults (Figure 3E). Interestingly, deconvolution analysis of the two samples that were excluded from the healthy children group because of the incidental detection of enterovirus and parainfluenza virus found that these samples had an increased percentage of neutrophils, thus supporting transcriptomic profiling of nasal mucosa as a sensitive tool for assessment of viral infections (Table E2). Taken together, these data demonstrate that samples obtained from the nasal mucosa of healthy and virus-infected participants are characterized by distinct epithelial- or immune-cell abundance profiles. Specifically, samples obtained from participants infected with SARS-CoV-2, IV, or RSV are characterized by an increased abundance of immune cells, whereas healthy participant samples are enriched for epithelial cells.
Figure 3.
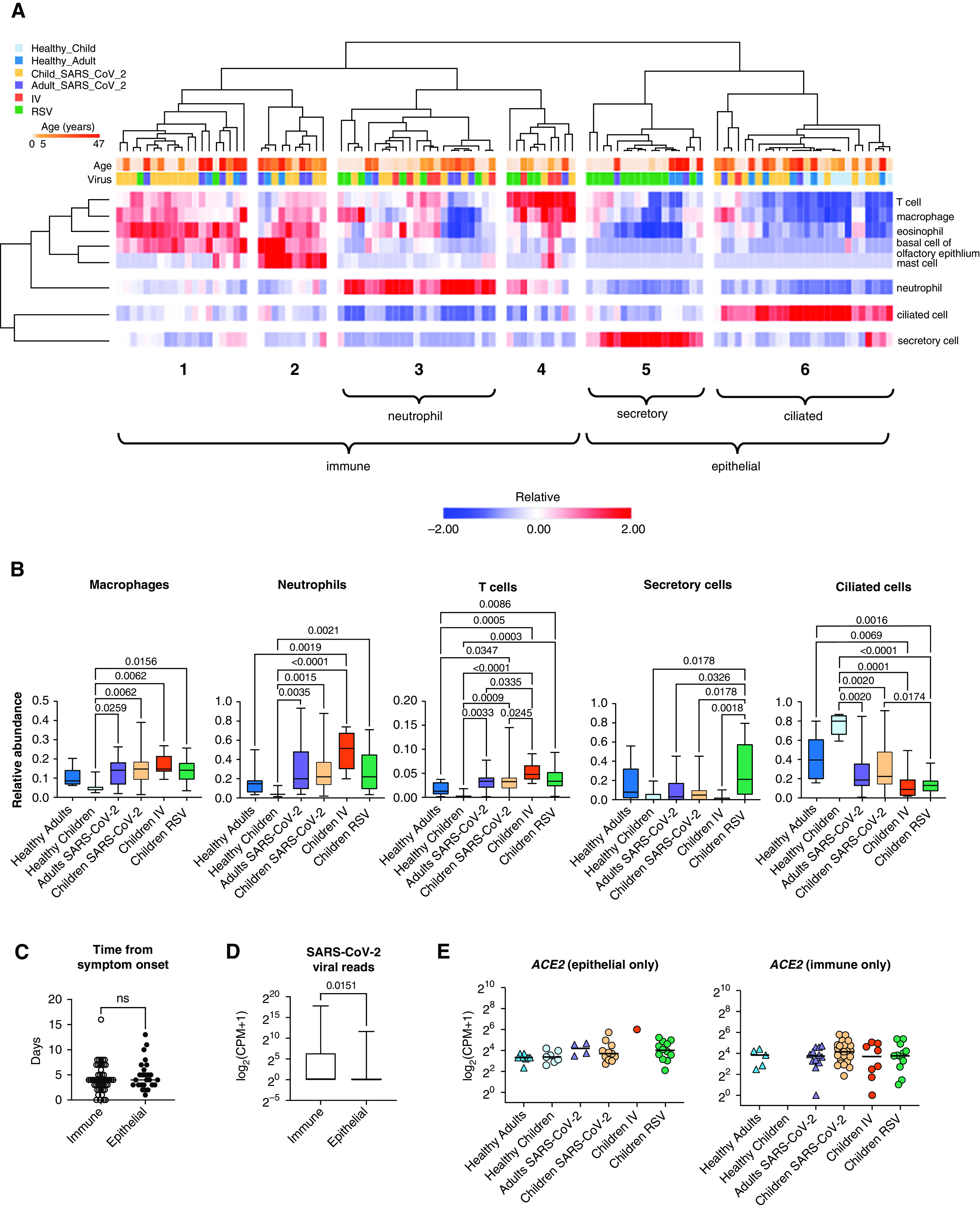
Cell-type deconvolution of the transcriptomic signatures in the nasal mucosa demonstrates an influx of immune cells after viral infection. (A) Heatmap demonstrating the relative abundance of cell types as determined by in silico deconvolution of bulk RNA-sequencing gene expression profile (see Methods). Hierarchical clustering on samples from 105 participants identified six clusters enriched for immune or epithelial cells. Z-score normalized relative abundance values are shown. (B) Relative abundance of immune and epithelial cell types across all six participant groups. Participants with a viral infection had a significantly higher relative abundance of immune cells than healthy participants. Adjusted P values calculated using the Kruskal-Wallis test with Benjamini-Hochberg FDR correction are shown. Adjusted P values < 0.05 were considered statistically significant. (C) Number of days between symptom onset and day of sample collection were similar for immune-enriched and epithelial-enriched samples. n.s. = not significant, by Mann-Whitney U test. (D) SARS-CoV-2 viral load, defined as average normalized reads for 11 SARS-CoV-2 viral genes, is significantly higher in immune-enriched clusters in comparison to epithelium-enriched clusters. Log-transformed pseudocounts (CPM + 1) are shown. The adjusted P value calculated by Mann-Whitney U test is shown. (E) Normalized expression of ACE2 in epithelial- and immune-enriched samples is similar across all participant groups; differences are not significant. Kruskal-Wallis testing with Benjamini-Hochberg FDR correction; all adjusted P values > 0.05. Log-transformed pseudocounts (CPM + 1) are shown.
Viral Infection Is Associated with Increased Expression of IFN and Innate Immune Response Genes in the Nasal Mucosa
To investigate whether specific gene expression patterns correlate with clinical parameters, we performed a WGCNA. Analysis of all participant samples (n = 105, both children and adults) identified seven modules, three of which significantly correlated with one or more clinical parameters (Figure 4A). Module 7 positively correlated with age and did not correlate with any other parameter, suggesting it contained age-related genes not associated with a viral infection. Enrichment analysis revealed that this age-related module enriched for genes involved in GO biological processes such as RNA processing and included several small nucleolar RNA genes (snoRNAs) such as SNORA49 and SNORD8 (Table E3), which have been proposed as aging markers (48) (Figure 4B). Modules 3 and 4 positively correlated with the number of SARS-CoV-2 viral reads. Module 3, which positively correlated with SARS-CoV-2 and IV reads, enriched for genes involved in response to virus and type I IFN signaling pathway (Figure 4B). Module 4 also correlated with RSV reads and enriched for genes involved in GO biological processes associated with neutrophil activation, chemotaxis, and leukocyte differentiation.
Figure 4.
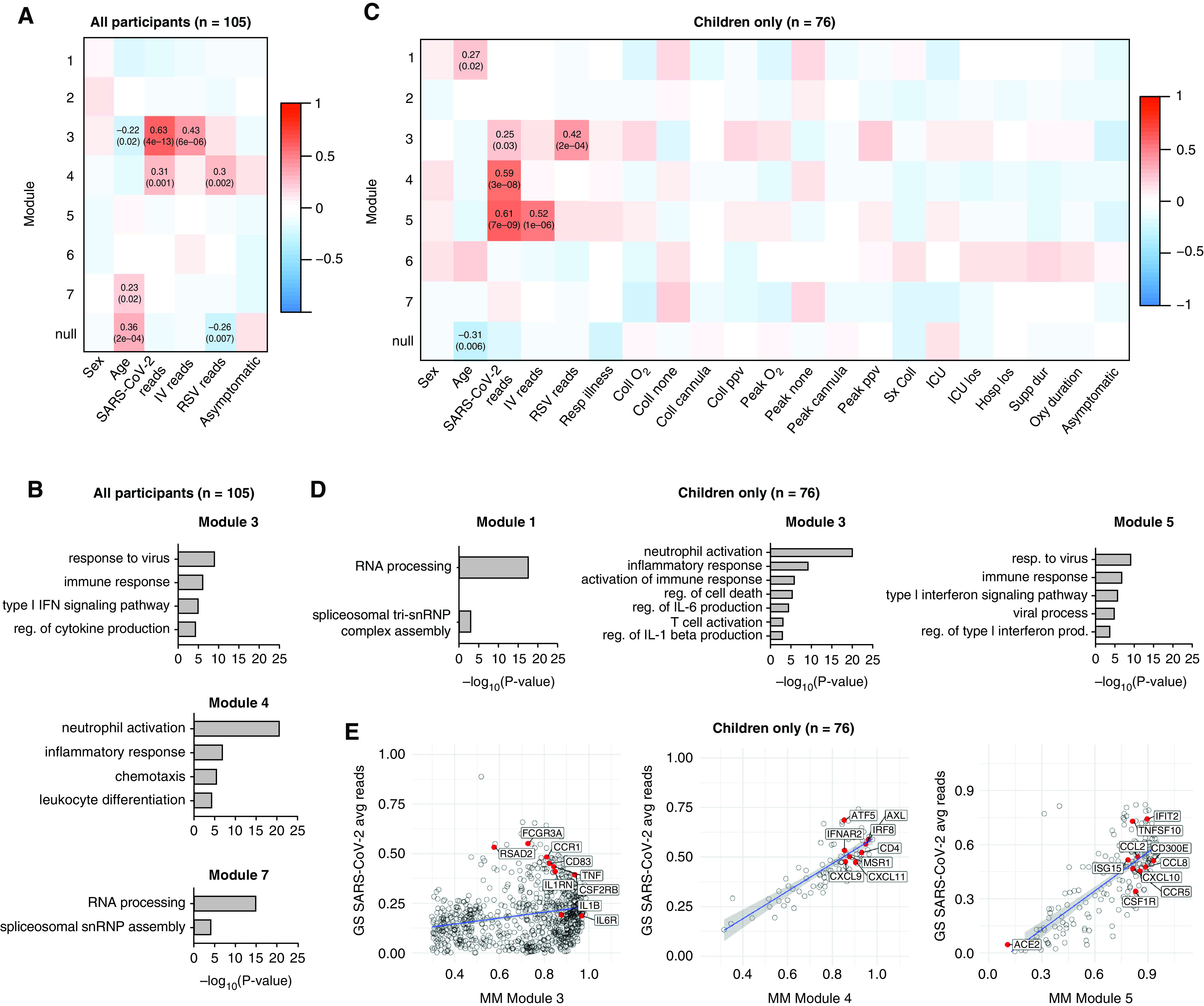
Transcriptional signatures associated with viral infection enrich for inflammatory and IFN response genes in children and adults. (A) A total of 2,170 HVGs for all participant samples (n = 105) were analyzed using WGCNA. Module–trait relationships were identified for seven modules. Modules 1–7 contain 33, 199, 248, 992, 375, 32, and 51 genes, respectively. A total of 240 genes that did not fall into any module were assigned to the null module. (B) Gene ontology (GO) biological process enrichment for modules correlating with SARS-CoV-2 viral reads. (C) A total of 2,024 HVGs identified among pediatric participant samples only (n = 76) were analyzed using WGCNA. Module–trait relationships were identified for seven modules. Modules 1–7 contain 43, 385, 823, 92, 200, 204 and 46 genes, respectively. A total of 231 genes that did not fall into any module were assigned to the null module. (D) GO biological process enrichment for modules correlating with SARS-CoV-2 reads. (E) Scatterplot of correlation between gene significance (GS) for SARS-CoV-2 reads versus module membership (MM) in modules 3, 4, and 5. Pearson correlation coefficients (r) and P values are shown. P values < 0.05 were considered statistically significant. Linear regression line (blue) with a 95% confidence interval (gray) are shown. Coll cannula = low-flow or high-flow nasal cannula at time of sample collection; Coll none = no respiratory support at time of sample collection; Coll O2 = supplemental oxygen at time of sample collection; Coll ppv = positive pressure ventilation (PPV) (noninvasive or invasive) at time of sample collection; Hosplos = hospital length of stay; HVGs = highly variable genes; ICU los = ICU length of stay; Oxy duration = duration of oxygen therapy; Peak cannula = low-flow or high-flow nasal cannula was the highest respiratory support received during hospitalization; Peak none = no respiratory support during hospitalization; Peak O2 = received supplemental oxygen during hospitalization; Peak ppv = PPV was highest respiratory support received during hospitalization; reg. = regulation; Resp Illness = respiratory illness; Supp dur = cumulative duration of any support (days); Sx coll = time between symptom onset and collection (days); WGCNA = weighted gene coexpression network analysis.
We next focused only on pediatric samples to identify distinct modules correlated with specific viral infections and clinical outcomes. WGCNA identified seven modules within the pediatric cohort (n = 76) (Figure 4C). Modules 3, 4, and 5 correlated with SARS-CoV-2 reads (Figure 4C). Module 3 also correlated with RSV reads and enriched for genes involved in the GO biological processes inflammatory response, regulation of IL-6 production, and T cell activation (Figure 4D). Module 5 correlated with both SARS-CoV-2 and IV reads and enriched for genes involved in response to virus, type I IFN signaling, and chemokines, including CXCL10 (Figure 4E). Module 4 was the only module unique to SARS-CoV-2 reads. The 92 genes that made up this module did not enrich for specific GO biological processes but included the antiviral transcription factor IRF8, the chemokines CXCL9 and CXCL11, and the T-cell marker CD4 (Figure 4E). There was no significant correlation between any of the seven modules and severity of illness or outcomes. Taken together, these data demonstrate that in participants with SARS-CoV-2, RSV, or IV infection, the nasal mucosa is characterized by the expression of genes associated with innate and adaptive immune responses.
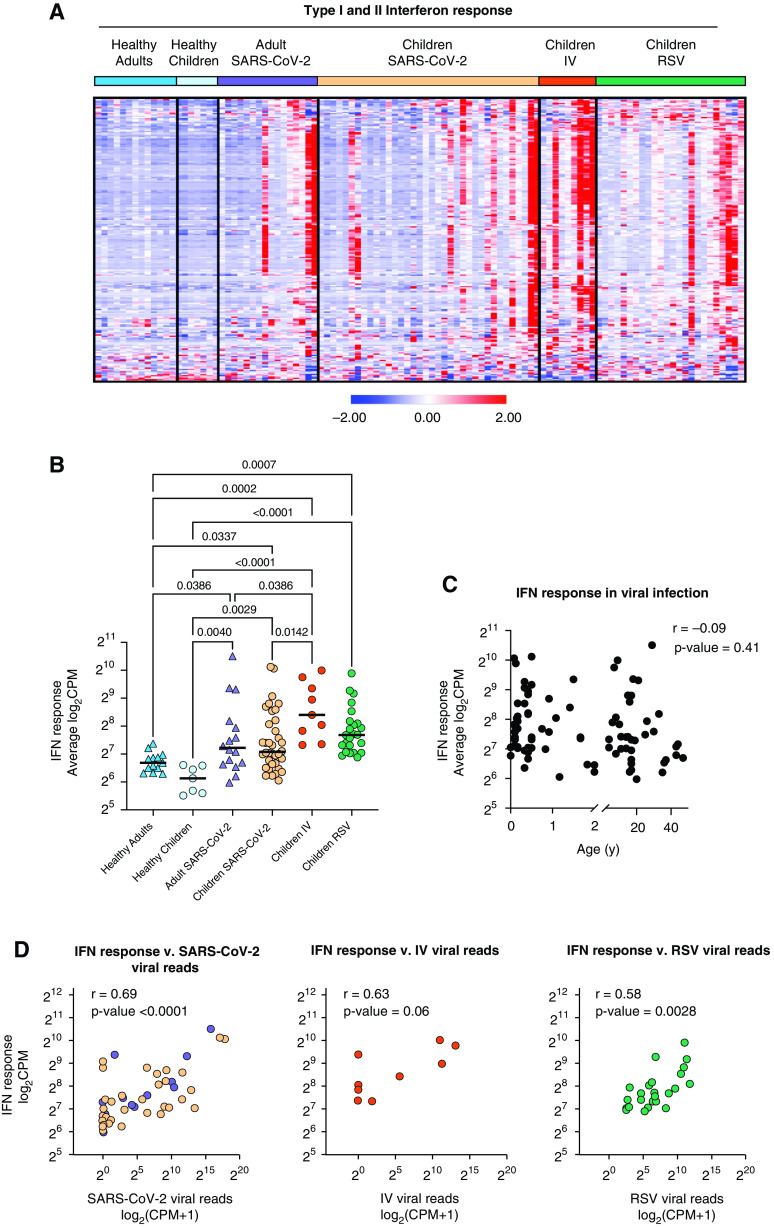
Local IFN Response Is Independent of Age and Virus
As WGCNA identified antiviral IFN signaling as a shared feature of viral infection with SARS-CoV-2, IV, or RSV in our cohort, we next focused on this pathway. We found that gene lists for type I and type II IFN responses largely overlap, with 75% of type I IFN response genes overlapping with type II IFN response genes. Therefore, we compiled a list of IFN-response genes using hallmark pathways for IFN-α and IFN-γ responses (49, 50). Comparison of normalized expression across all participant groups revealed an increased expression of IFN response genes in patients with RSV, IV, and SARS-CoV-2 compared with healthy controls that did not correlate with age (Figures 5A–5C and Table E4). Specifically, we did not observe differences in the IFN response between children and adults infected with SARS-CoV-2 (Figure 5B). Although neither age nor type of viral infection correlated with the IFN response, we found a significant correlation with the viral load measured by an average number of viral reads (Figure 5D). Consistent with the lack of correlation between viral load, the severity of illness, or the time from symptom onset, the IFN response did not correlate with clinical outcomes such as duration of respiratory support and ICU length of stay (Figures E3A and E3B). Interestingly, although we detected an IFN response signature in the majority of virus-infected participants, we did not detect the expression of secreted type I, type II, or type III IFNs at the transcriptional level in a majority of patients with RSV or SARS-CoV-2 infection (Figure 5E and Table E4). Secreted IFNs were primarily detected in participants with IV infection, consistent with this cohort having the highest average expression of IFN response genes overall (Figures 5B and 5E). This finding suggests that the nasal mucosa is not a source of secreted IFNs in RSV and SARS-CoV-2 infection.
Figure 5.
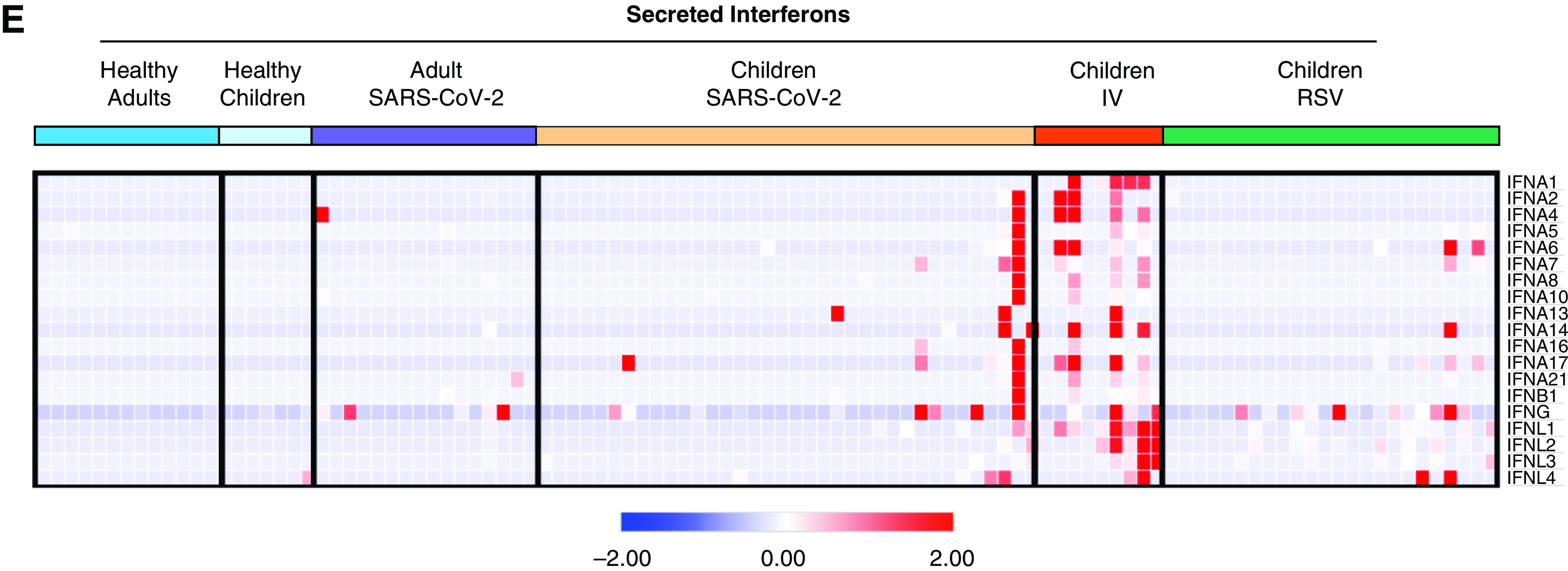
Local IFN response is independent of age and virus. (A) Heatmap of z-score normalized expression (CPM) of 222 type I and II IFN response genes. Hierarchical clustering on genes (rows) did not identify distinct clusters. Samples within SARS-CoV-2, IV, and RSV groups are ordered by ascending viral reads for SARS-CoV-2, IV, or RSV, respectively. (B) Average normalized expression of IFN response genes (combined Hallmark pathway lists for IFN-α and IFN-γ response) by participant group. (Adjusted P value < 0.05 is considered statistically significant, Kruskal-Wallis test with Benjamini-Hochberg FDR correction). Log-transformed CPM counts are shown. (C) Scatter plot of correlation between age (years) and average IFN response reads. Log-transformed CPM counts are shown. Pearson correlation coefficient (r) and P value are shown. (D) Scatter plot of correlation between average IFN response reads (log2CPM) and SARS-CoV-2, RSV, or IV viral reads, respectively. Pearson r values and P values are shown. (E) Heatmap of z-score normalized expression (CPM) for 19 secreted type I, II, and III IFNs.
Adults, but Not Children, with Asymptomatic to Mild Disease Upregulate T-Cell Activation, TCR Signaling, and Neutrophil Activation Genes
As the IFN response was not different between children and adults infected with SARS-CoV-2 (Figure 5B), we sought to determine how the expression of genes in other immune-related pathways compared in children and adults with asymptomatic to mild SARS-CoV-2 infection. We focused on participants with high viral loads (average SARS-CoV-2 viral read > 6 CPM, median threshold within our cohort) to ensure active SARS-CoV-2 infection (Figure E4A). In line with our previous observation among all participants (Figure 5B), no difference in the magnitude of the IFN response between this subset of children and adults was observed (Figure E4A). The cellular composition was also similar between groups, allowing us to compare gene expression differences without cellular composition bias (Figure E4B). We did not observe differences in ACE2 or TMPRSS2 expression between children and adults (Figure E4A). The comparison of children and adults with asymptomatic to mild disease and high SARS-CoV-2 viral load identified 737 differentially expressed genes (Figure 6A). Genes upregulated in children compared with adults (IL18, IL18RAP, IL23A, and KRT17) were enriched for epithelial repair processes (keratinization and regulation of epithelium regeneration) and distinct innate immune response-related processes (response to TNF and regulation of leukocyte activation) (Figures 6B and 6C and Table E5). This observation was supported by additional enrichment analyses, demonstrating keratinization and TNF-signaling gene sets were significantly enriched in children compared with adults (Figure E4C). In line with our previous findings in Figure 5 and Figure E4A, GSEA demonstrated that genes involved in the IFN response were not significantly enriched in children or adults with high viral load and asymptomatic to mild SARS-CoV-2 infection (Figure E4D). Instead, genes upregulated in adults compared with children enriched for distinct innate immune response processes involved in neutrophil function and chemotaxis (neutrophil function and IL-8 signaling) (Figures 6B and 6C). Expression of neutrophil chemotactic factor IL8 and its receptors, CXCR1 and CXCR2, were upregulated in adults compared with children (Figure 6C). Of note, IL17RA, the receptor for the inflammatory cytokine IL-17, which is secreted by activated T cells (51, 52), was upregulated in adults compared with children (Figure 6C). These data suggest that T-cell signaling and activation are upregulated in adults compared with children. To further investigate whether transcriptional signatures in adults enriched for T-cell function, we performed GSEA and found that genes involved in TCR signaling and T-cell activation (CD3E, PTEN, ITK, VAV1, and HLA-E) were significantly enriched in adults compared with children (Figure 6D).
Figure 6.
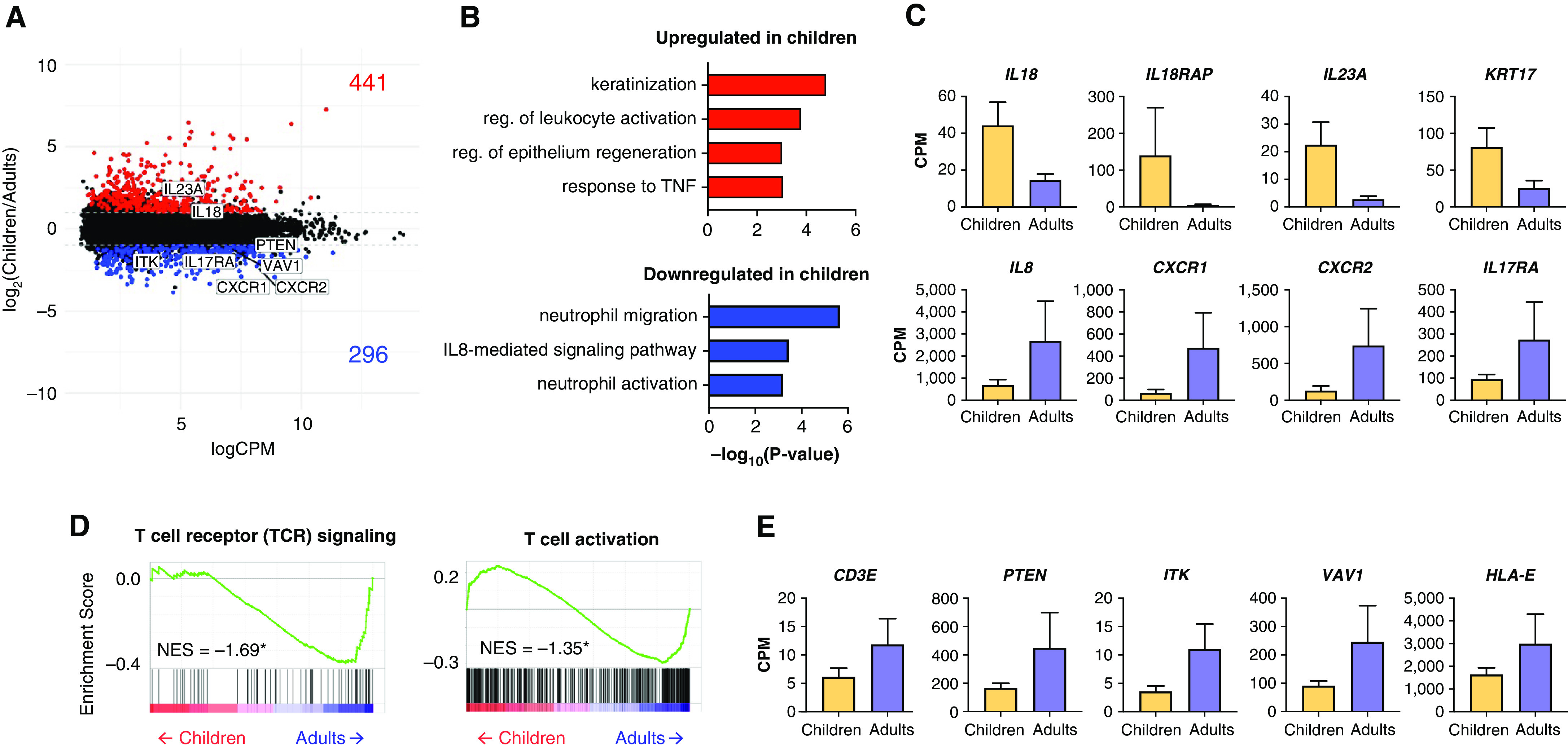
Adults, but not children, with asymptomatic to mild disease upregulate TCR signaling, T-cell activation, and neutrophil activation genes with equivalent viral loads (A) MA plot of differentially expressed genes (edgeR P value < 0.05 and log2FC = |1.0|) between adults and children with mild, asymptomatic disease and high SARS-CoV-2 viral load (>6 CPM average). Numbers of genes upregulated (red) and downregulated (blue) in children compared with adults are shown. (B) Genes upregulated in children (red) enriched for keratinization and epithelial repair GO biological processes. Genes upregulated in adults (blue) enriched for neutrophil-related GO biological processes. (C) Bar graphs of gene expression (CPM) for representative up- and downregulated genes in children compared with adults are shown. All differentially expressed genes shown passed the cutoff as defined in the MA plot in panel (A): edgeR P value < 0.05 and log2FC > |1.0|, and are statistically significant. Error bars represent SEM. (D) Gene set enrichment analysis enrichment plots demonstrating TCR signaling and T-cell activation is negatively enriched in children compared with adults. Gene set enrichment analysis was performed on gene lists ranked by edgeR reported fold-change between children and adults with mild, asymptomatic disease and high SARS-CoV-2 viral load (>6 CPM average). Normalized enrichment scores (NES) are shown. *Statistically significant FDR values < 0.05. (E) Bar graphs of gene expression (CPM) for representative leading-edge genes, upregulated in adults, are shown. Error bars represent SEM.
Taken together, these data suggest that, compared with children, the immune response in the nasal mucosa of SARS-CoV-2–infected adults with active asymptomatic to mild disease is characterized by increased TCR signaling and downstream T-cell activation.
Discussion
The majority of children infected with SARS-CoV-2 experience mild symptoms similar to the common cold. The mechanisms underlying this relative protection from severe infection in children compared with adults are unclear. Differences in expression of the SARS-CoV-2 entry factors or response to the virus at the primary site of infection between children and adults have been proposed as potential mechanisms (53, 54); however, these hypotheses have not been directly tested to date. In our cohort of children infected with SARS-CoV-2, the local IFN response in the nasal mucosa was largely the same as non-hospitalized adults infected with SARS-CoV-2 and children with severe RSV infection. This similarity in IFN responses across age, virus, and severity of illness suggests that age-related differences in SARS-CoV-2 infection outcomes are independent of the primary IFN response to the virus in the nasal mucosa.
Age-related differences in expression of ACE2 and TMPRSS2 have been proposed as modulators of clinical severity of SARS-CoV-2 infections. ACE2 is the cell surface receptor bound by the SARS-CoV-2 spike protein, and TMPRSS2 is a host protease that cleaves the spike protein to allow for efficient binding and cell entry (8). Previous studies have demonstrated an association between age and the expression of ACE2 and TMPRSS2, suggesting that low levels of expression of ACE2 and TMPRSS2 in pediatric airway epithelial cells could protect children from severe infection (11, 12, 40). In our cohort, we did not observe a difference in the expression of ACE2 or TMPRSS2 between children and adults, in agreement with recent publications (55, 56). Although some reports suggested that ACE2 expression may increase in response to IFN stimulation (7, 40), we saw no increase in the expression of ACE2 and TMPRSS2 expression during active SARS-CoV-2 or other viral infections, supporting previous reports that IFN signaling does not lead to increased ACE2 expression (41–43).
Our study’s unique strength is the inclusion of the appropriate controls: healthy children and adults and children infected with other respiratory viruses. This allowed us to report our findings on SARS-CoV-2 infection in the context of RSV and IV infections, which are known to induce severe illness requiring hospitalization and respiratory support. We found a high degree of similarity in the local response to SARS-CoV-2, RSV, and IV infections. This finding agrees with Mick and colleagues’ report using metatranscriptomics on nasopharyngeal swabs that revealed substantial overlap between SARS-CoV-2 and other acute respiratory infections in adults (57). Although there was one module of 92 genes that only correlated with SARS-CoV-2 viral reads and not RSV or IV viral reads, this module did not correlate with severity of illness or outcomes. Thus, our findings suggest that other factors, such as systemic immune responses, may determine the differences in disease severity between children with SARS-CoV-2, RSV, and IV.
Given that the expression of SARS-CoV-2 entry factors and the local IFN response at the primary site of infection does not differ between children and adults, other factors must mediate the relative protection in children and unpredictable susceptibility in elderly adults infected with SARS-CoV-2. We observed an age-related difference in neutrophil and T-cell–related gene expression in participants with asymptomatic to mild SARS-CoV-2 infection. Despite similar relative abundance of neutrophils in the nasal mucosa (Figure 3), adults with SARS-CoV-2 infection demonstrated increased expression of neutrophil migration and activation genes compared with children. The biological impact of the increased gene expression in adults, but not children, is unclear. Future studies sampling the nasal mucosa over several days would provide useful insight into the functional consequence of these differences in gene expression on neutrophil recruitment and activation over the course of infection. In a study exposing healthy adult volunteers to RSV, the presence of neutrophilic inflammatory signals in the nasal mucosa before exposure was associated with development of symptomatic disease (58). This suggests neutrophil activation may be a risk factor for symptomatic viral infection in adults. Children with severe viral pneumonia have been reported to have an increased ratio of CD8+ to CD4+ T cells (59). We observed that the relative abundance of T cells in the nasal mucosa was similar in our cohort of children and adults infected with SARS-CoV-2. However, CD8 was a leading-edge gene in the T cell activation GSEA, which was enriched in adults compared with children. In contrast, CD4 had one of the highest enrichment scores in the TCR pathway GSEA, which was enriched in children compared with adults (Figure 6D, Figure E4E, and Table E6). These exploratory data suggest a potential age-related difference in T-cell subset ratios in the nasal mucosa of children and adults infected with SARS-CoV-2. Further investigation is needed to assess how differences in neutrophil activation and T-cell subsets may contribute to age-related differences in severity of SARS-CoV-2 infection.
Even before the COVID-19 pandemic, a substantial number of individuals in the population had antibodies and T cells that recognize β coronaviruses and can cross-react with SARS-CoV-2. These antibodies and T cells are generated over time as individuals are exposed to seasonal coronaviruses (53, 54). In our cohort, we observed an increase in TCR signaling and T-cell activation gene expression in adults compared with children with asymptomatic to mild SARS-CoV-2. This finding may represent increased TCR signaling and activation in the setting of cross-reactive T cells. Such cross-reactive memory T cells have been reported for SARS-CoV (60) and recently for SARS-CoV-2 (61–65). This observation initially led to speculation that these cross-reactive antibodies and T cells could play a protective role in children and young adults and that vanishing adaptive immunity would make elderly individuals more susceptible to SARS-CoV-2. However, in another study, a larger number of SARS-CoV-2 cross-reactive memory T cells was observed in the elderly and patients with severe compared with mild COVID-19 (65). Importantly, these cross-reactive T cells exhibited lower avidity and reduced antiviral responses when stimulated with SARS-CoV-2 peptides compared with T cells from patients who recovered from COVID-19. Thus, instead of being protective, these cross-reactive, low-avidity T cells, which are less efficient at virus elimination, could contribute to prolonged inflammatory signaling. This hypothesis is attractive as it explains the differences in the epidemiology of SARS-CoV-2, which disproportionately affects elderly individuals. Children and young adults, who likely have fewer lifetime coronavirus exposures, often have only mild symptoms, despite having viral loads in the upper airways comparable to adults (3, 14). Although multiple studies have investigated the adaptive immune response in adults infected with COVID-19, similar systematic studies in the pediatric population are lacking.
Our study has several limitations. First, our institution used two different PCR platforms for the detection of SARS-CoV-2 during the study period. Owing to differences in methodology, viral loads cannot be directly compared between the platforms. Moreover, PCR testing was not available for the adults accompanying children infected with SARS-CoV-2 during the study period. To overcome this limitation, we used dual RNA-seq, which allows for simultaneous host transcriptomic analysis and pathogen characterization (21). By creating custom hybrid genomes, we were able to identify and quantify viral transcripts in our samples as a surrogate for viral load, thus enabling comparison across all groups in our study. Second, although our study includes children infected with other respiratory viruses (RSV and IV), which serve as proper controls to SARS-CoV-2–infected children, it primarily includes hospitalized children with SARS-CoV-2, representing a minority of pediatric SARS-CoV-2 infections. However, 14% of our pediatric cohort were asymptomatic with incidental SARS-CoV-2 diagnoses and 71% of symptomatic SARS-CoV-2–infected children never required supplemental oxygen or other respiratory support, making them comparable to our adult cohort. The local host response in the nasal mucosa of asymptomatic children could not be distinguished from more severely affected pediatric participants, suggesting factors outside the upper respiratory tract drive severe SARS-CoV-2 infections in children. Third, our transcriptomic analysis was performed on a biopsy from a complex tissue containing multiple epithelial and immune cell types (44). Thus, our results would have been affected by the changes in the abundance of the specific cell types, either owing to age- or disease-related differences in tissue composition or operator sampling bias, if we did not account for sample composition. We applied in silico tissue deconvolution (38) to account for differences in tissue composition, which allowed us to perform a more accurate comparison between samples with similar compositions. This comparison demonstrated that among subjects with high SARS-CoV-2 viral load and similar cellular composition, children exhibit decreased TCR signaling, decreased T cell activation, and increased macrophage activation on a per-cell level. The use of complementary techniques that enable analysis at the single-cell level, such as single-cell RNA-seq, will allow for a more cell-type–specific assessment of changes associated with SARS-CoV-2 or other viral infections. Recently, single-cell RNA-seq has been applied to understand virus-host interactions in children infected with IV (66), and similar efforts are underway in SARS-CoV-2 (67). However, single-cell RNA-seq has several limitations that do not apply to bulk RNA-seq. Sample processing and cryopreservation are tolerated to different degrees by different cell types, with certain populations, such as neutrophils, particularly difficult to recover. Unlike with bulk RNA-seq, where cells are immediately lysed and stored for RNA isolation, true cell proportions will not be captured using cryopreserved samples for single-cell RNA-seq.
Conclusions
We present evidence that the IFN response to SARS-CoV-2 infection at the primary site of infection is similar between children and adults and comparable in magnitude to other common respiratory viruses. In contrast, TCR signaling, T-cell activation, and neutrophil activation is increased in adults compared with children with mild SARS-CoV-2 infection (Figure E5). Our findings suggest that adults may be at increased risk for severe SARS-CoV-2 infection compared with children owing to age-related differences in the cellular immune response to SARS-CoV-2.
Acknowledgments
Acknowledgments
The authors thank the Genomics Compute Cluster, which is jointly supported by the Feinberg School of Medicine, the Center for Genetic Medicine, and Feinberg's Department of Biochemistry and Molecular Genetics, the Office of the Provost, the Office for Research, and Northwestern Information Technology. The Genomics Compute Cluster is part of Quest, Northwestern University's high-performance computing facility, to advance research in genomics. Figure E5 was created using BioRender.com. They also thank all the participants for enrolling in this study. The authors thank Rogan Grant for his help with the RNA-seq pipeline and G.R.S. Budinger for their thoughtful discussions.
Footnotes
Supported by the Gorter Family Foundation (A.D.P.); National Institutes of Health grants U19AI135964, P01AG049665, and R01HL153312, and Northwestern University Clinical and Translational Sciences Institute COVID-19 Rapid Response Grant (A.V.M.); National Institutes of Health grants P01AG049665 and P01GM096971 (K.M.R.); Stanley Manne Research Institute COVID-19 Springboard Exploratory Research Award, American Thoracic Society Unrestricted Grant: Pulmonary, and National Institutes of Health grant K08HL143127 (B.M.C.); National Institutes of Health grants R01HL149883, R01HL153122, and R01HL114800 (B.D.S.); and the Stanley Manne Research Institute COVID-19 Springboard Exploratory Research Award (T.H.-S. and L.N.S.-P.).
Author Contributions: C.M.K., A.D.P., A.S., L.N.S.-P., W.J.M., A.V.M., K.M.R., and B.M.C. contributed to the acquisition, analysis, and interpretation of the data and drafting the manuscript. L.S., J.C., N.S.M., and G.R.H. contributed to the acquisition and interpretation of the data. K.R.A., H.C.D.U., H.A.-V., and Y.P. contributed to the acquisition and analysis of the data. C.M.K., A.D.P., G.R.H., T.H.-S., L.N.S.-P., W.J.M., B.D.S., A.V.M., K.M.R., and B.M.C. contributed to the conception and design of the study and the acquisition and interpretation of the data. All authors critically revised and approved the final version of the manuscript.
This article has a data supplement, which is accessible from this issue’s table of contents at www.atsjournals.org.
Originally Published in Press as DOI: 10.1165/rcmb.2021-0292OC on November 3, 2021
Author disclosures are available with the text of this article at www.atsjournals.org.
References
- 1. Castagnoli R, Votto M, Licari A, Brambilla I, Bruno R, Perlini S, et al. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection in children and adolescents: a systematic review. JAMA Pediatr . 2020;174:882–889. doi: 10.1001/jamapediatrics.2020.1467. [DOI] [PubMed] [Google Scholar]
- 2. Ludvigsson JF. Systematic review of COVID-19 in children shows milder cases and a better prognosis than adults. Acta Paediatr . 2020;109:1088–1095. doi: 10.1111/apa.15270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Dong Y, Mo X, Hu Y, Qi X, Jiang F, Jiang Z, et al. Epidemiology of COVID-19 among children in China. Pediatrics . 2020;145:e20200702. doi: 10.1542/peds.2020-0702. [DOI] [PubMed] [Google Scholar]
- 4.COVID-19 cases, deaths, and trends in the US Centers for Disease Control and Prevention; 2020. [updated 28 Mar 2020; accessed 29 Dec 2020]. Available from: https://covid.cdc.gov/covid-data-tracker/ [Google Scholar]
- 5. Leeb RT, Price S, Sliwa S, Kimball A, Szucs L, Caruso E, et al. COVID-19 trends among school-aged children - United States, March 1-September 19, 2020. MMWR Morb Mortal Wkly Rep . 2020;69:1410–1415. doi: 10.15585/mmwr.mm6939e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Sungnak W, Huang N, Bécavin C, Berg M, Queen R, Litvinukova M, et al. HCA Lung Biological Network SARS-CoV-2 entry factors are highly expressed in nasal epithelial cells together with innate immune genes. Nat Med . 2020;26:681–687. doi: 10.1038/s41591-020-0868-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ziegler CGK, Allon SJ, Nyquist SK, Mbano IM, Miao VN, Tzouanas CN, et al. HCA Lung Biological Network SARS-CoV-2 receptor ACE2 is an IFN-stimulated gene in human airway epithelial cells and is detected in specific cell subsets across tissues. Cell . 2020;181:1016–1035.e19. doi: 10.1016/j.cell.2020.04.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hoffmann M, Kleine-Weber H, Schroeder S, Krüger N, Herrler T, Erichsen S, et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell . 2020;181:271–280.e8. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Walls AC, Park Y-J, Tortorici MA, Wall A, McGuire AT, Veesler D. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell . 2020;183:1735. doi: 10.1016/j.cell.2020.11.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wrapp D, Wang N, Corbett KS, Goldsmith JA, Hsieh C-L, Abiona O, et al. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science . 2020;367:1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bunyavanich S, Do A, Vicencio A. Nasal gene expression of angiotensin-converting enzyme 2 in children and adults. JAMA . 2020;323:2427–2429. doi: 10.1001/jama.2020.8707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Muus C, Luecken MD, Eraslan G, Sikkema L, Waghray A, Heimberg G, et al. Single-cell meta-analysis of SARS-CoV-2 entry genes across tissues and demographics. Nat Med . 2021;27:546–559. doi: 10.1038/s41591-020-01227-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Yonker LM, Neilan AM, Bartsch Y, Patel AB, Regan J, Arya P, et al. Pediatric severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2): clinical presentation, infectivity, and immune responses. J Pediatr . 2020;227:45–52.e5. doi: 10.1016/j.jpeds.2020.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Jones TC, Biele G, Muhlemann B, Veith T, Schneider J, Beheim-Schwarzbach J, et al. Estimating infectiousness throughout SARS-CoV-2 infection course. Science . 2021;373:eabi5273. doi: 10.1126/science.abi5273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Heald-Sargent T, Muller WJ, Zheng X, Rippe J, Patel AB, Kociolek LK. Age-related differences in nasopharyngeal severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) levels in patients with mild to moderate coronavirus disease 2019 (COVID-19) JAMA Pediatr . 2020;174:902–903. doi: 10.1001/jamapediatrics.2020.3651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Baggio S, L’Huillier AG, Yerly S, Bellon M, Wagner N, Rohr M, et al. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) viral load in the upper respiratory tract of children and adults with early acute coronavirus disease 2019 (COVID-19) Clin Infect Dis . 2021;73:148–150. doi: 10.1093/cid/ciaa1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Iwasaki A, Foxman EF, Molony RD. Early local immune defences in the respiratory tract. Nat Rev Immunol . 2017;17:7–20. doi: 10.1038/nri.2016.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Blanco-Melo D, Nilsson-Payant BE, Liu W-C, Uhl S, Hoagland D, Møller R, et al. Imbalanced host response to SARS-CoV-2 drives development of COVID-19. Cell . 2020;181:1036–1045.e9. doi: 10.1016/j.cell.2020.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hadjadj J, Yatim N, Barnabei L, Corneau A, Boussier J, Smith N, et al. Impaired type I IFN activity and inflammatory responses in severe COVID-19 patients. Science . 2020;369:718–724. doi: 10.1126/science.abc6027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Do LAH, Pellet J, van Doorn HR, Tran AT, Nguyen BH, Tran TTL, et al. Host transcription profile in nasal epithelium and whole blood of hospitalized children under 2 years of age with respiratory syncytial virus infection. J Infect Dis . 2017;217:134–146. doi: 10.1093/infdis/jix519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wesolowska-Andersen A, Everman JL, Davidson R, Rios C, Herrin R, Eng C, et al. Dual RNA-seq reveals viral infections in asthmatic children without respiratory illness which are associated with changes in the airway transcriptome. Genome Biol . 2017; 18:12. doi: 10.1186/s13059-016-1140-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Poole A, Urbanek C, Eng C, Schageman J, Jacobson S, O’Connor BP, et al. Dissecting childhood asthma with nasal transcriptomics distinguishes subphenotypes of disease. J Allergy Clin Immunol . 2014;133:670–8.e12. doi: 10.1016/j.jaci.2013.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Altman MC, Gill MA, Whalen E, Babineau DC, Shao B, Liu AH, et al. Transcriptome networks identify mechanisms of viral and nonviral asthma exacerbations in children. Nat Immunol . 2019;20:637–651. doi: 10.1038/s41590-019-0347-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Symptoms of COVID-19. Centers for Disease Control and Prevention; 2021. [updated 19 May 2020; accessed 24 Jun 2021]. Available from: https://www.cdc.gov/coronavirus/2019-ncov/symptoms-testing/symptoms.html [Google Scholar]
- 25. Harris PA, Taylor R, Thielke R, Payne J, Gonzalez N, Conde JG. Research electronic data capture (REDCap)--a metadata-driven methodology and workflow process for providing translational research informatics support. J Biomed Inform . 2009;42:377–381. doi: 10.1016/j.jbi.2008.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Pediatric Acute Lung Injury Consensus Conference Group. Pediatric acute respiratory distress syndrome: consensus recommendations from the Pediatric Acute Lung Injury Consensus Conference. Pediatr Crit Care Med . 2015;16:428–439. doi: 10.1097/PCC.0000000000000350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Sala MA, Balderas-Martínez YI, Buendía-Roldan I, Abdala-Valencia H, Nam K, Jain M, et al. Inflammatory pathways are upregulated in the nasal epithelium in patients with idiopathic pulmonary fibrosis. Respir Res . 2018;19:233. doi: 10.1186/s12931-018-0932-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Di Tommaso P, Chatzou M, Floden EW, Barja PP, Palumbo E, Notredame C. Nextflow enables reproducible computational workflows. Nat Biotechnol . 2017;35:316–319. doi: 10.1038/nbt.3820. [DOI] [PubMed] [Google Scholar]
- 29. Ewels PA, Peltzer A, Fillinger S, Patel H, Alneberg J, Wilm A, et al. The nf-core framework for community-curated bioinformatics pipelines. Nat Biotechnol . 2020;38:276–278. doi: 10.1038/s41587-020-0439-x. [DOI] [PubMed] [Google Scholar]
- 30. Kurtzer GM, Sochat V, Bauer MW. Singularity: scientific containers for mobility of compute. PLoS One . 2017;12:e0177459. doi: 10.1371/journal.pone.0177459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics . 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Liao Y, Smyth GK, Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics . 2014;30:923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 33. Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics . 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. McCarthy DJ, Chen Y, Smyth GK. Differential expression analysis of multifactor RNA-seq experiments with respect to biological variation. Nucleic Acids Res . 2012;40:4288–4297. doi: 10.1093/nar/gks042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Eden E, Navon R, Steinfeld I, Lipson D, Yakhini Z. GOrilla: a tool for discovery and visualization of enriched GO terms in ranked gene lists. BMC Bioinformatics . 2009;10:48. doi: 10.1186/1471-2105-10-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics . 2008;9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Mootha VK, Lindgren CM, Eriksson K-F, Subramanian A, Sihag S, Lehar J, et al. PGC-1α-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet . 2003;34:267–273. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- 38. Aliee H, Theis FJ. AutoGeneS: Automatic gene selection using multi-objective optimization for RNA-seq deconvolution. Cell Syst . 2021;12:706–715.e704. doi: 10.1016/j.cels.2021.05.006. [DOI] [PubMed] [Google Scholar]
- 39. Ordovas-Montanes J, Dwyer DF, Nyquist SK, Buchheit KM, Vukovic M, Deb C, et al. Allergic inflammatory memory in human respiratory epithelial progenitor cells. Nature . 2018;560:649–654. doi: 10.1038/s41586-018-0449-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Sajuthi SP, DeFord P, Li Y, Jackson ND, Montgomery MT, Everman JL, et al. Type 2 and IFN inflammation regulate SARS-CoV-2 entry factor expression in the airway epithelium. Nat Commun . 2020;11:5139. doi: 10.1038/s41467-020-18781-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Chu H, Chan JF-W, Wang Y, Yuen TT-T, Chai Y, Hou Y, et al. Comparative replication and immune activation profiles of SARS-CoV-2 and SARS-CoV in human lungs: an ex vivo study with implications for the pathogenesis of COVID-19. Clin Infect Dis . 2020;71:1400–1409. doi: 10.1093/cid/ciaa410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Onabajo OO, Banday AR, Stanifer ML, Yan W, Obajemu A, Santer DM, et al. Interferons and viruses induce a novel truncated ACE2 isoform and not the full-length SARS-CoV-2 receptor. Nat Genet . 2020;52:1283–1293. doi: 10.1038/s41588-020-00731-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. de Lang A, Osterhaus ADME, Haagmans BL. Interferon-gamma and interleukin-4 downregulate expression of the SARS coronavirus receptor ACE2 in Vero E6 cells. Virology . 2006;353:474–481. doi: 10.1016/j.virol.2006.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Deprez M, Zaragosi L-E, Truchi M, Becavin C, Ruiz García S, Arguel M-J, et al. A single-cell atlas of the human healthy airways. Am J Respir Crit Care Med . 2020;202:1636–1645. doi: 10.1164/rccm.201911-2199OC. [DOI] [PubMed] [Google Scholar]
- 45. Vieira Braga FA, Kar G, Berg M, Carpaij OA, Polanski K, Simon LM, et al. A cellular census of human lungs identifies novel cell states in health and in asthma. Nat Med . 2019;25:1153–1163. doi: 10.1038/s41591-019-0468-5. [DOI] [PubMed] [Google Scholar]
- 46. Persson BD, Jaffe AB, Fearns R, Danahay H. Respiratory syncytial virus can infect basal cells and alter human airway epithelial differentiation. PLoS One . 2014;9:e102368. doi: 10.1371/journal.pone.0102368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Sebina I, Phipps S. The contribution of neutrophils to the pathogenesis of RSV bronchiolitis. Viruses . 2020;12:E808. doi: 10.3390/v12080808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Steinbusch MMF, Fang Y, Milner PI, Clegg PD, Young DA, Welting TJM, et al. Serum snoRNAs as biomarkers for joint ageing and post traumatic osteoarthritis. Sci Rep . 2017;7:43558. doi: 10.1038/srep43558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA . 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Liberzon A, Subramanian A, Pinchback R, Thorvaldsdóttir H, Tamayo P, Mesirov JP. Molecular signatures database (MSigDB) 3.0. Bioinformatics . 2011;27:1739–1740. doi: 10.1093/bioinformatics/btr260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. McKenzie DR, Kara EE, Bastow CR, Tyllis TS, Fenix KA, Gregor CE, et al. IL-17-producing γδ T cells switch migratory patterns between resting and activated states. Nat Commun . 2017;8:15632. doi: 10.1038/ncomms15632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Nakae S, Saijo S, Horai R, Sudo K, Mori S, Iwakura Y. IL-17 production from activated T cells is required for the spontaneous development of destructive arthritis in mice deficient in IL-1 receptor antagonist. Proc Natl Acad Sci USA . 2003;100:5986–5990. doi: 10.1073/pnas.1035999100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Steinman JB, Lum FM, Ho PP-K, Kaminski N, Steinman L. Reduced development of COVID-19 in children reveals molecular checkpoints gating pathogenesis illuminating potential therapeutics. Proc Natl Acad Sci USA . 2020;117:24620–24626. doi: 10.1073/pnas.2012358117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zimmermann P, Curtis N.Why is COVID-19 less severe in children? A review of the proposed mechanisms underlying the age-related difference in severity of SARS-CoV-2 infections Arch Dis Child [online ahead of print] 1 Dec 2020 10.1136/archdischild-2020-320338 [DOI] [PubMed] [Google Scholar]
- 55.Loske J, Röhmel J, Lukassen S, Stricker S, Magalhães VG, Liebig J, et al. Pre-activated antiviral innate immunity in the upper airways controls early SARS-CoV-2 infection in children Nat Biotechnol [online ahead of print] 18 Aug 2021 10.1038/s41587-021-01037-9 [DOI] [PubMed] [Google Scholar]
- 56. Pierce CA, Sy S, Galen B, Goldstein DY, Orner E, Keller MJ, et al. Natural mucosal barriers and COVID-19 in children. JCI Insight . 2021;6:148694. doi: 10.1172/jci.insight.148694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Mick E, Kamm J, Pisco AO, Ratnasiri K, Babik JM, Castañeda G, et al. Upper airway gene expression reveals suppressed immune responses to SARS-CoV-2 compared with other respiratory viruses. Nat Commun . 2020;11:5854. doi: 10.1038/s41467-020-19587-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Habibi MS, Thwaites RS, Chang M, Jozwik A, Paras A, Kirsebom F, et al. Neutrophilic inflammation in the respiratory mucosa predisposes to RSV infection. Science . 2020;370:eaba9301. doi: 10.1126/science.aba9301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Connors TJ, Ravindranath TM, Bickham KL, Gordon CL, Zhang F, Levin B, et al. Airway CD8(+) T cells are associated with lung injury during infant viral respiratory tract infection. Am J Respir Cell Mol Biol . 2016;54:822–830. doi: 10.1165/rcmb.2015-0297OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Gioia C, Horejsh D, Agrati C, Martini F, Capobianchi MR, Ippolito G, et al. T-cell response profiling to biological threat agents including the SARS coronavirus. Int J Immunopathol Pharmacol . 2005;18:525–530. doi: 10.1177/039463200501800312. [DOI] [PubMed] [Google Scholar]
- 61. Grifoni A, Weiskopf D, Ramirez SI, Mateus J, Dan JM, Moderbacher CR, et al. Targets of T cell responses to SARS-CoV-2 coronavirus in humans with COVID-19 disease and unexposed individuals. Cell . 2020;181:1489–1501.e15. doi: 10.1016/j.cell.2020.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Le Bert N, Tan AT, Kunasegaran K, Tham CYL, Hafezi M, Chia A, et al. SARS-CoV-2-specific T cell immunity in cases of COVID-19 and SARS, and uninfected controls. Nature . 2020;584:457–462. doi: 10.1038/s41586-020-2550-z. [DOI] [PubMed] [Google Scholar]
- 63. Mateus J, Grifoni A, Tarke A, Sidney J, Ramirez SI, Dan JM, et al. Selective and cross-reactive SARS-CoV-2 T cell epitopes in unexposed humans. Science . 2020;370:89–94. doi: 10.1126/science.abd3871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Braun J, Loyal L, Frentsch M, Wendisch D, Georg P, Kurth F, et al. SARS-CoV-2-reactive T cells in healthy donors and patients with COVID-19. Nature . 2020;587:270–274. doi: 10.1038/s41586-020-2598-9. [DOI] [PubMed] [Google Scholar]
- 65. Bacher P, Rosati E, Esser D, Martini GR, Saggau C, Schiminsky E, et al. Low-avidity CD4+ T cell responses to SARS-CoV-2 in unexposed individuals and humans with severe COVID-19. Immunity . 2020;53:1258–1271.e5. doi: 10.1016/j.immuni.2020.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cao Y, Guo Z, Vangala P, Donnard E, Liu P, McDonel P, et al. Single-cell analysis of upper airway cells reveals host-viral dynamics in influenza infected adults bioRxiv;2020. [accessed 2021 Jun 3]. Available from: https://www.biorxiv.org/content/10.1101/2020.04.15.042978v1.
- 67.Ballestar E, Farber DL, Glover S, Horwitz B, Meyer K, Nikolić M, et al. Chan Zuckerberg Initiative Single-Cell COVID-19 Consortia Single cell profiling of COVID-19 patients: an international data resource from multiple tissues bioRxiv 2020. [accessed 2021 Jun 3]. Available from: https://www.medrxiv.org/content/10.1101/2020.11.20.20227355v1.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Processed RNA-seq counts are available as Table E1. Raw data is in the process of being deposited to dbGaP/SRA. Code is available via https://github.com/NUPulmonary/2021_Koch.