Figure 4.
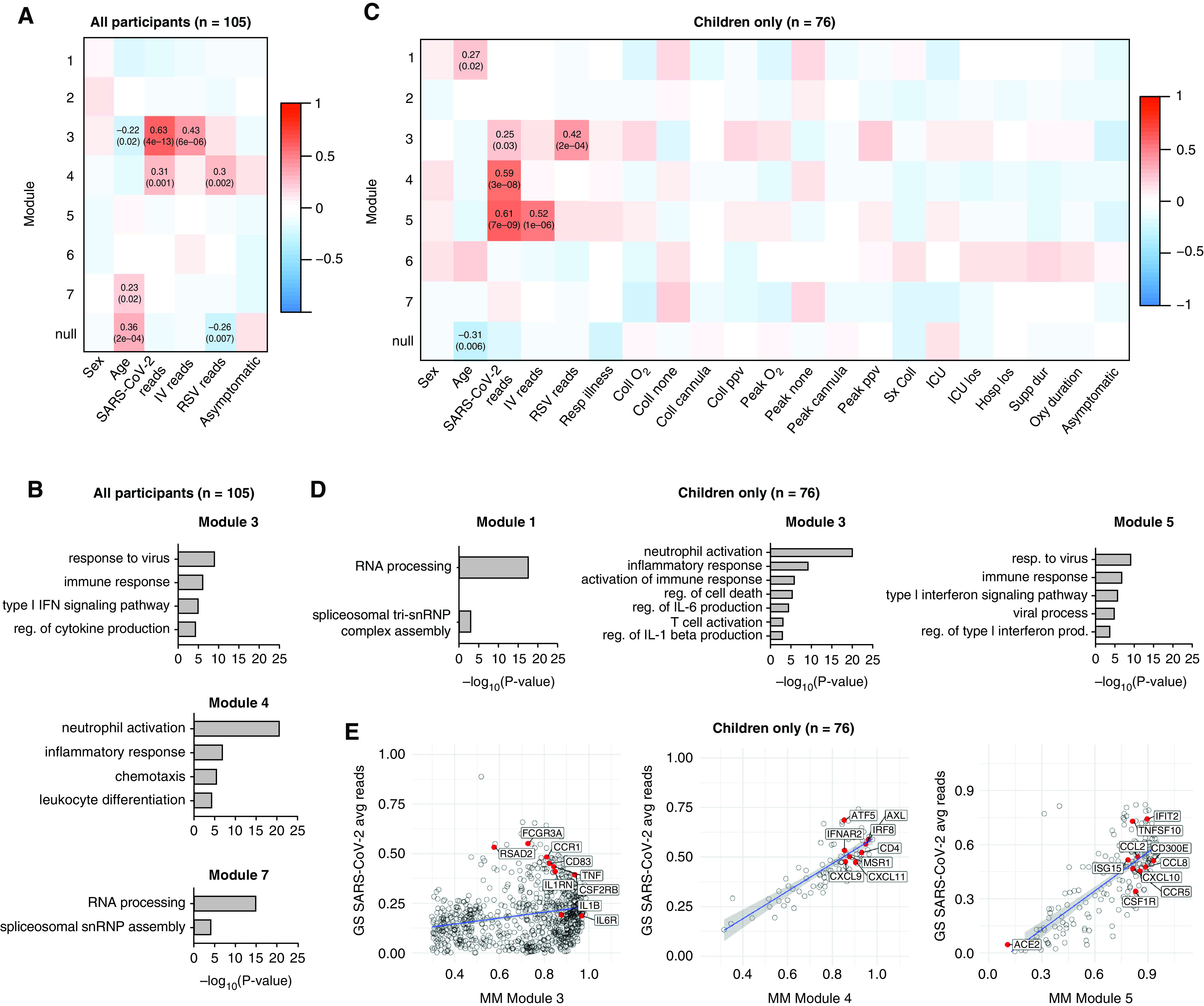
Transcriptional signatures associated with viral infection enrich for inflammatory and IFN response genes in children and adults. (A) A total of 2,170 HVGs for all participant samples (n = 105) were analyzed using WGCNA. Module–trait relationships were identified for seven modules. Modules 1–7 contain 33, 199, 248, 992, 375, 32, and 51 genes, respectively. A total of 240 genes that did not fall into any module were assigned to the null module. (B) Gene ontology (GO) biological process enrichment for modules correlating with SARS-CoV-2 viral reads. (C) A total of 2,024 HVGs identified among pediatric participant samples only (n = 76) were analyzed using WGCNA. Module–trait relationships were identified for seven modules. Modules 1–7 contain 43, 385, 823, 92, 200, 204 and 46 genes, respectively. A total of 231 genes that did not fall into any module were assigned to the null module. (D) GO biological process enrichment for modules correlating with SARS-CoV-2 reads. (E) Scatterplot of correlation between gene significance (GS) for SARS-CoV-2 reads versus module membership (MM) in modules 3, 4, and 5. Pearson correlation coefficients (r) and P values are shown. P values < 0.05 were considered statistically significant. Linear regression line (blue) with a 95% confidence interval (gray) are shown. Coll cannula = low-flow or high-flow nasal cannula at time of sample collection; Coll none = no respiratory support at time of sample collection; Coll O2 = supplemental oxygen at time of sample collection; Coll ppv = positive pressure ventilation (PPV) (noninvasive or invasive) at time of sample collection; Hosplos = hospital length of stay; HVGs = highly variable genes; ICU los = ICU length of stay; Oxy duration = duration of oxygen therapy; Peak cannula = low-flow or high-flow nasal cannula was the highest respiratory support received during hospitalization; Peak none = no respiratory support during hospitalization; Peak O2 = received supplemental oxygen during hospitalization; Peak ppv = PPV was highest respiratory support received during hospitalization; reg. = regulation; Resp Illness = respiratory illness; Supp dur = cumulative duration of any support (days); Sx coll = time between symptom onset and collection (days); WGCNA = weighted gene coexpression network analysis.
