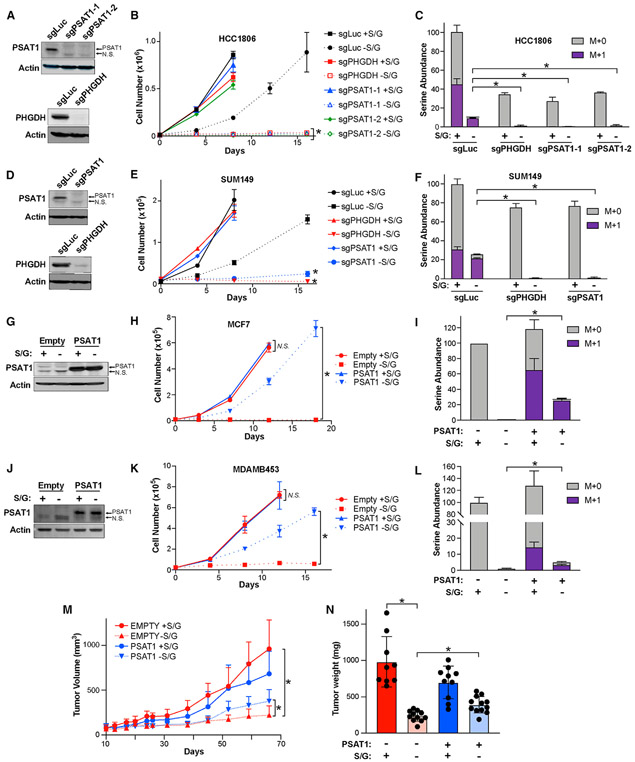
Figure 3. PSAT1 expression determines growth in the absence of exogenous serine.
(A) Representative western blots of control (sgLuc), PSAT1 knockout (sgPSAT1-1 and sgPSAT1-2), and PHGDH knockout (sgPHGDH) HCC1806 cells.
(B) Growth curve of control (sgLuc), sgPHGDH, and sgPSAT1 HCC1806 cells grown with or without S/G. Values are the means ± SDs of triplicate samples from an experiment representative of 3 independent experiments. *p < 0.05 in 2-way repeated measures ANOVA. sgPSAT1 or sgPHGDH without S/G data were compared to sgLuc without S/G data.
(C) Serine abundance and biosynthesis in sgLuc, sgPHGDH, and sgPSAT1 HCC1806 cells treated with or without S/G for 48 h. M + 1 (in purple) indicates “heavy” serine made in the serine synthesis pathway. Values are the means ± SDs of triplicate samples from an experiment representative of 2 independent experiments. *p < 0.05 from unpaired 2-sided t tests on serine abundance data.
(D) Representative western blots of control (sgLuc), PSAT1 knockout (sgPSAT1), and PHGDH knockout (sgPHGDH) SUM149 cells.
(E) Growth curve of control (sgLuc), sgPHGDH, and sgPSAT1 SUM149 cells grown with or without S/G. Values are the means ± SDs of triplicate samples from an experiment representative of 3 independent experiments. *p < 0.05 in 2-way repeated measures ANOVA. sgPSAT1 or sgPHGDH without S/G data were compared to sgLuc without S/G data.
(F) Serine abundance and biosynthesis in sgLuc, sgPHGDH, and sgPSAT1 SUM149 cells treated with or without S/G for 48 h. M + 1 (in purple) indicates “heavy” serine made in the serine synthesis pathway. Values are the means ± SDs of triplicate samples from an experiment representative of 2 independent experiments. *p < 0.05 from unpaired 2-sided t tests on serine abundance data.
(G) Representative western blot of control (Empty) and PSAT1 overexpressing MCF7 cells (PSAT1) cultured with or without S/G for 48 h.
(H) Growth curve of Empty and PSAT1 MCF7 cells cultured with or without S/G. Values are the means ± SDs of triplicate samples from an experiment representative of 3 independent experiments. *p < 0.05 and N.S. indicates p > 0.05 in 2-way repeated measures ANOVA tests comparing Empty and PSAT1 cells either with or without S/G.
(I) Serine abundance and biosynthesis in Empty and PSAT1 MCF7 cells cultured with or without S/G for 48 h. M + 1 (in purple) indicates “heavy” serine made in the serine synthesis pathway. Values are the means ± SDs of triplicate samples from an experiment representative of 2 independent experiments. *p < 0.05 from unpaired 2-sided t tests on serine abundance data.
(J) Representative western blot of control (Empty) and PSAT1 overexpressing MDAMB453 cells (PSAT1) with or without S/G for 48 h.
(K) Growth curve of Empty and PSAT1 MDAMB453 cells cultured with or without S/G. Values are the means ± SDs of triplicate samples from an experiment representative of 3 independent experiments. *p < 0.05 and N.S. indicates p > 0.05 in 2-way repeated-measures ANOVA tests comparing Empty and PSAT1 cells either with or without S/G.
(L) Serine abundance and biosynthesis in Empty and PSAT1 MDAMB453 cells cultured with or without S/G for 48 h. M + 1 (in purple) indicates “heavy” serine made in the serine synthesis pathway. Values are the means ± SDs of triplicate samples from an experiment representative of 2 independent experiments. *p < 0.05 from unpaired 2-sided t tests on serine abundance data.
(M) Tumor volume over time after injecting Empty and PSAT1 MCF7 cells into the mammary gland of the nude mice fed with or without S/G diet. Values are the means ± SDs (n = 9 tumors for Empty with S/G, n = 11 tumors for Empty without S/G, n = 10 tumors for PSAT1 with S/G and n = 12 tumors for PSAT1 without S/G). *p < 0.05 in mixed-effects model analyses.
(N) Empty and PSAT1 MCF7 xenograft tumor weight as measured at endpoint. Values are the means ± SDs (n = 9 tumors for Empty with S/G, n = 11 tumors for Empty without S/G, n = 10 tumors for PSAT1 with S/G and n = 12 tumors for PSAT1 without S/G). *p < 0.05 in 2-sided Welch’s t tests.