FIG 3.
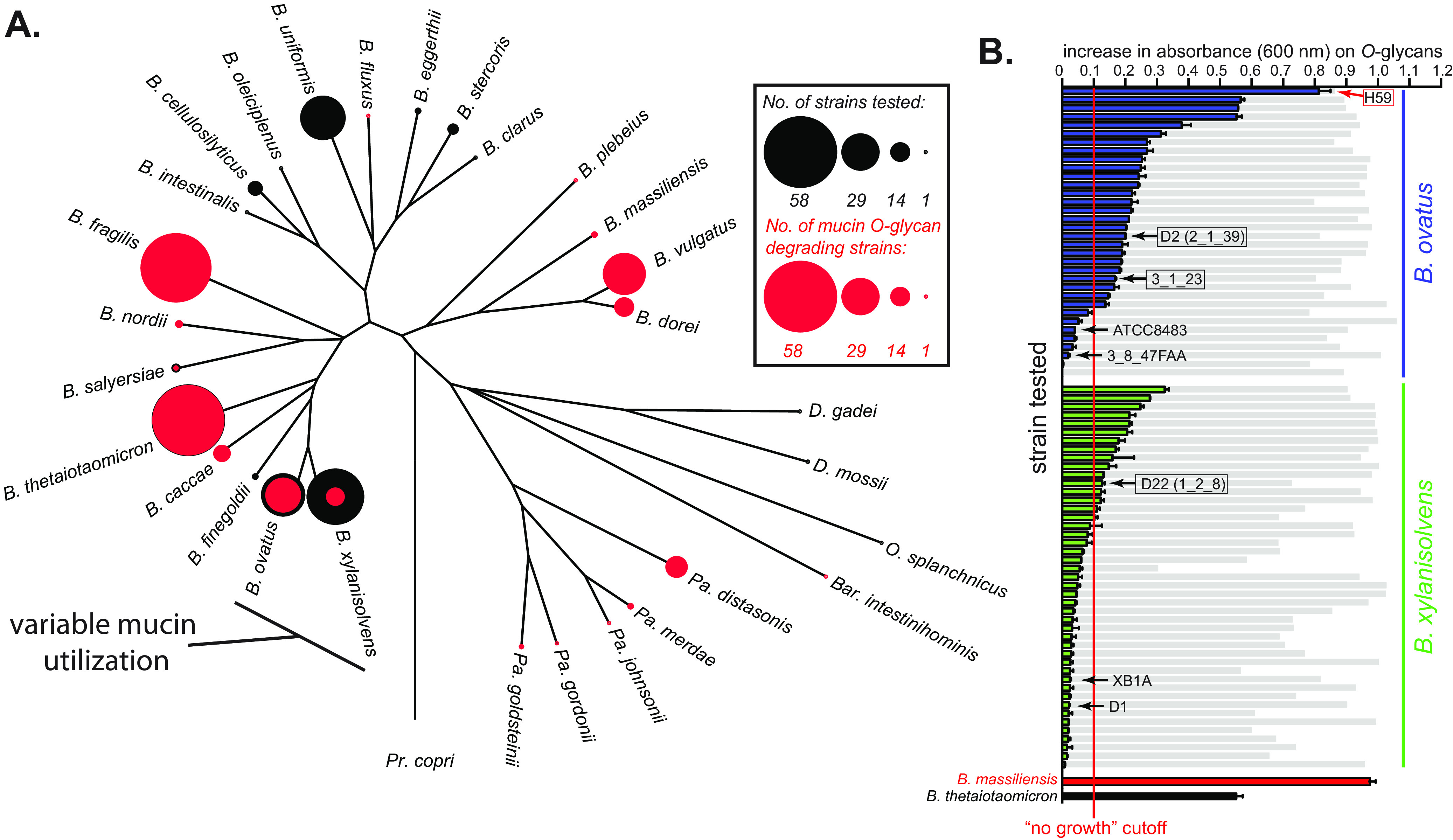
Host mucin O-glycan metabolism within the Bacteroides. (A) A phylogenetic tree based on housekeeping genes that compares mucin O-glycan utilization across species. The diameter of the black circles represents the number of strains tested within each species (sample depth), whereas the size of the overlaid red circle corresponds to the number of strains exhibiting O-glycan metabolism. Note that some species have either full or no penetrance of this phenotypic trait and yet others like B. ovatus/B. xylanisolvens have more extensive variability among strains. (B) Strains of B. ovatus (blue) and B. xylanisolvens (green) that show variable growth abilities on mucin O-glycan (n = 2 growth assays per bar, error bars are range between values). Gray histogram bars are total growth controls on an aggregate of the monosaccharides that all strains of these two species grow on (Table S1) and are provided as a reference for overall growth ability on a non O-glycan substrate. Data from two established O-glycan degraders, namely, B. massiliensis and B. thetaiotaomicron, are also shown for reference. Species with black arrows were used for pangenome analyses to compare genetic traits associated with mucin O-glycan metabolism. We performed RNA-seq on three strains included in this pangenome analysis (black boxes) that were positive for O-glycan utilization and an additional strain, namely, B. ovatus NLAE-zl-H59 (red arrow, box), to see if there were unique genes/PULs present in strains that have the ability to grow on mucin O-glycans.
