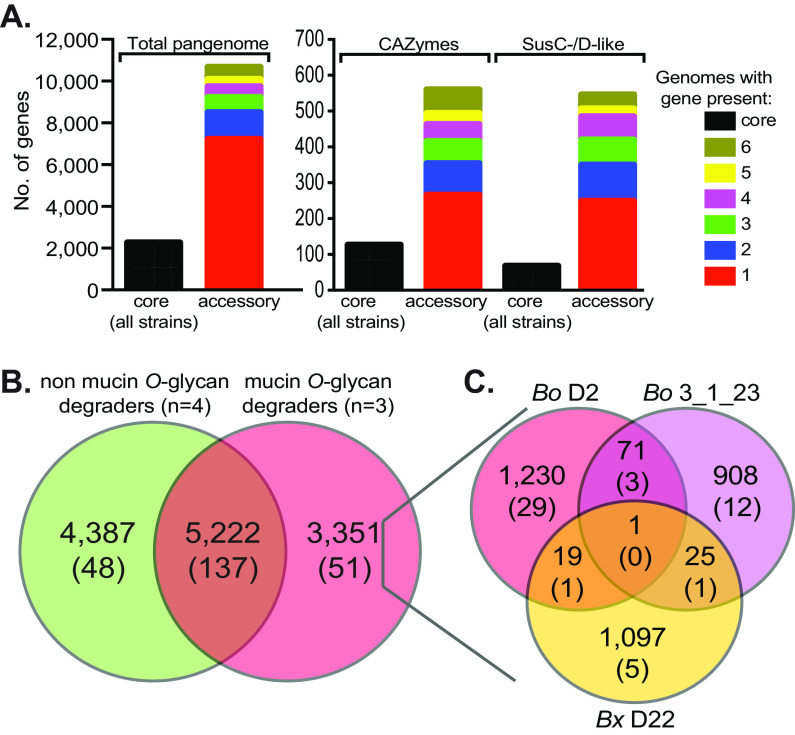
FIG 4.
Distribution of all genes as well as core polysaccharide utilization functions in the B. ovatus/B. xylanisolvens pangenome. (A) Left, shows the number of core genes (i.e., those present in all 7 strains used for pangenome construction) compared with genes present in 2 to 7 of the individual strains. Right, shows the same distribution of genes assigned to PULs or particular degradative CAZyme families (GH, PL, and CE) (see Tables S2 and S3 for more detailed assignments). (B) The distribution of genes between mucin-degrading (n = 3) and nondegrading (n = 4) strains used to construct the pangenome. Top numbers indicate total genes, while numbers in parentheses indicate the number of PULs (not individual PUL genes) in each category. (C) Distribution of the genes that are unique to the three mucin-degrading strains within each genome. Genes/PULs are numbered as described for panel B. Note that no PULs are shared by all three strains.