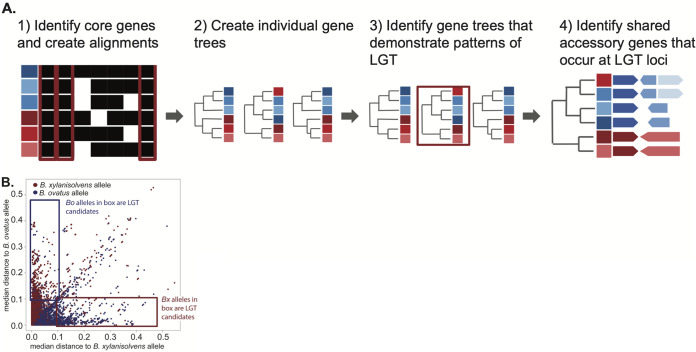
FIG 6.
Identification of putative lateral gene transfer events. (A) Schematic of the workflow to identify putative LGT core genes, which is described as follows: align genes and build corresponding trees for each core gene, determine the median substitution distances for each allele of a core gene in a given strain to both species, and identify loci with an identical conserved structure between isolates of opposite species. (B) Plot of median distances for all core genes identified in the 33 genomes analyzed. The boxes show the regions containing genes for which the median distance was >0.1 to the assigned species for a given strain and ≤0.1 for the opposite species to which a strain is assigned. These genes were determined to be high-confidence examples of core/node genes that had been replaced by an allele from the other species.