Figure 2. DC-LC3in binds to LC3B and covalently modifies Lysine 49.
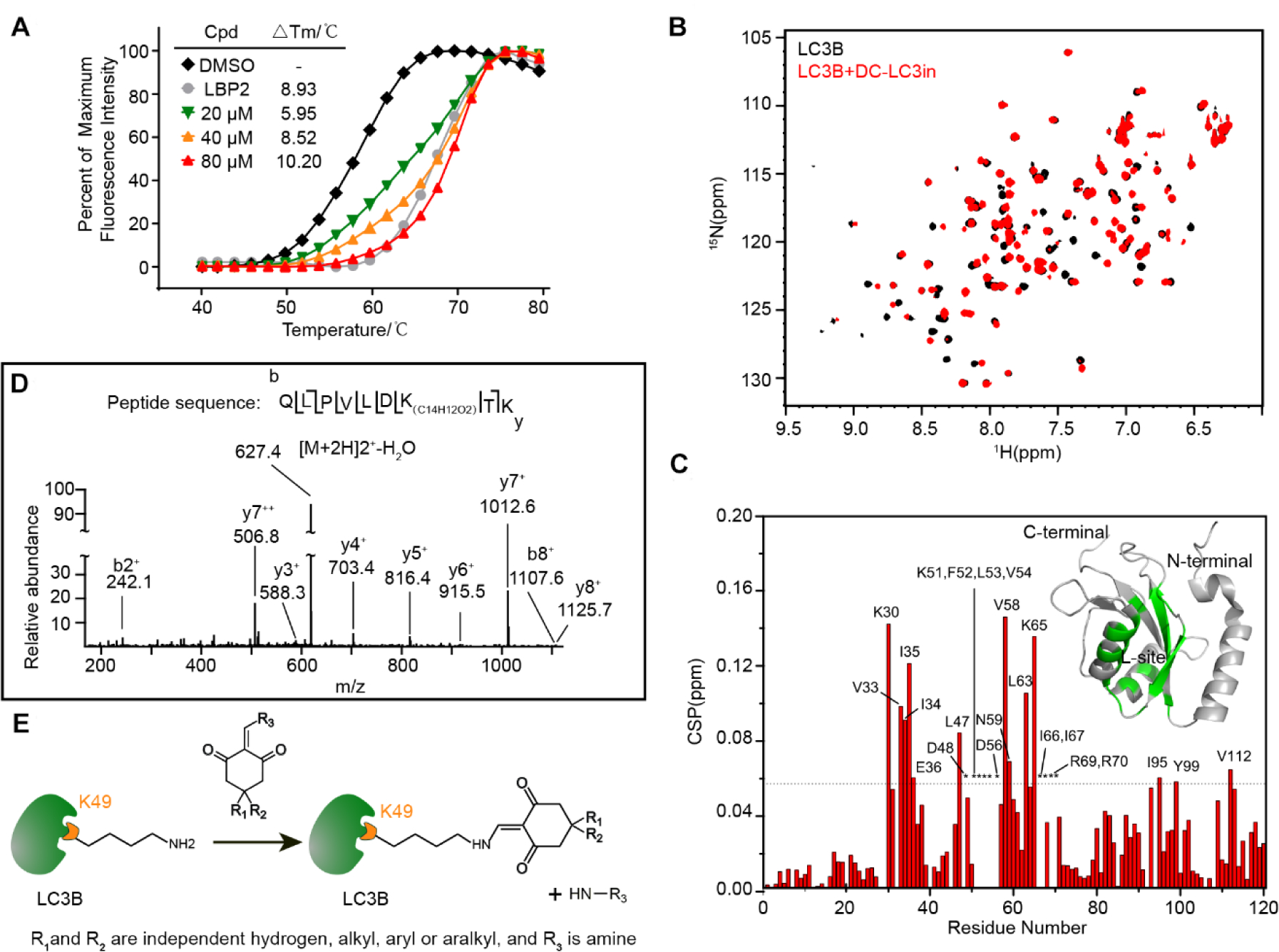
(A) Change in thermodynamic stability of LC3B upon binding with DC-LC3in and LBP2. The △Tm was calculated as the difference vs. the DMSO control sample. (B) Overlapped 1H-15N HSQC spectra for LC3B in the presence (red) or absence (black) of DC-LC3in at a 1:1 probe:protein ratio. (C) Chemical shift perturbation (CSP) analysis for LC3B DC-LC3in binding, and structural mapping of LC3B based on the CSP data. Residues with CSP values over the mean ± SD (dashed line) or those with attenuated resonance signals (indicated by asterisk) are labeled. Residues perturbed significantly or attenuated upon the binding of DC-LC3in are colored (green) on a 3D cartoon structure of LC3B (PDB ID 1V49). (D) HPLC-nESI MS/MS scan of the fragmented precursor ion of an LC3B peptide (m/z = 627.4), demonstrating that LC3B was covalently modified by DC-LC3in at Lysine 49. The difference in the detected peptide fragment mass vs. the theoretically predicted m/z for modified peptide corresponds to the monoisotopic mass for a C14H12O2 isomer. (E) Proposed model for the reaction between DC-LC3in analogs and LC3B’s Lys49 residue.
