FIG 3.
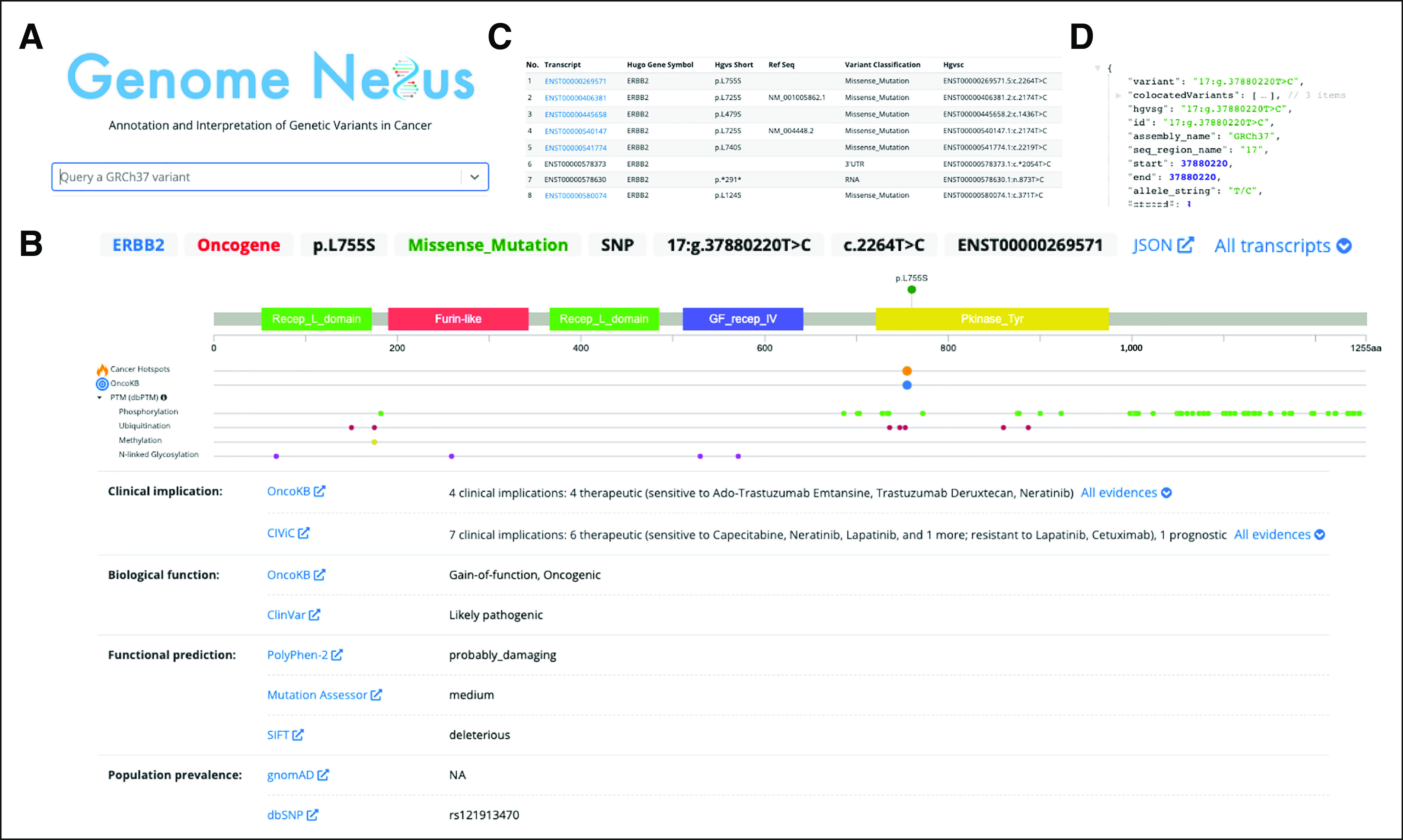
Interpretation of a single cancer variant using Genome Nexus. The ERBB2 L755S variant is used as an example. The homepage allows one to search for (A) a variant, which then leads the user to (B) the Variant Page. This page includes a lollipop diagram, showing the domains (PFAM) and annotations from several other resources (Cancer Hotspots, OncoKB, dbPTM, Uniprot, and variant effect predictor). Each of these annotation sources is hyperlinked when hovering and can be investigated in more detail. Below the lollipop plot, there is a summary of information for each annotation source. They are grouped by higher-level categories, such as clinical implication, biologic function, functional prediction, and population prevalence. The displayed transcript is ENST00000269571. One can investigate any transcript for ERBB2 by clicking on (C) the All transcripts dropdown. The raw data from the application programming interface can be seen in (D). CIViC, Clinical Interpretation of Variants in Cancer; dbPTM, database for protein Post-Translational Modifications; dbSNP, database for Single Nucleotide Polymorphisms; NA, not applicable; OncoKB, Oncology Knowledge Base; PTM, Post-Translational Modification; SIFT, Sorting Tolerant From Intolerant.
