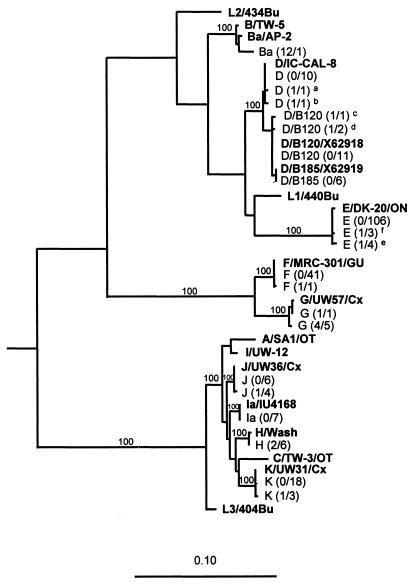
FIG. 1.
Phylogenetic tree showing the evolutionary relationship between the C. trachomatis nucleotide sequences of the omp1 gene from one representative of each genetic variant in the clinical material and 18 serovars of C. trachomatis available from GenBank. Sequences representing C. psittaci, C. pneumoniae, and a murine variant of C. trachomatis (MoPn) were used as outgroups to root the tree. The tree was calculated by the maximum-likelihood method (transition/transversion ratio, 1.4; global rearrangement option on) from an alignment of 1,032 aligned nucleotides. Relevant bootstrap values (as a percentage of 1,000 replicates) are also given. Reference strains are shown in boldface text. The clinical strains (n = 237) are illustrated with genotype and number of mutations per number of samples (within parentheses). Footnote letters: a, genotype D with one mutation at position 331 compared to D/IC-CAL-8; b, genotype D with one mutation at position 1092 compared to D/IC-CAL-8; c, genotype D/B120 with one mutation at position 975 compared to the strain X62918 (GenBank); d, genotype D/B120 with one mutation at position 1045 compared to the strain X62918 (GenBank); e, genotype E with one mutation at position 420 compared to E/DK20/ON; f, genotype E with one mutation at position 997 compared to E/DK20/ON.