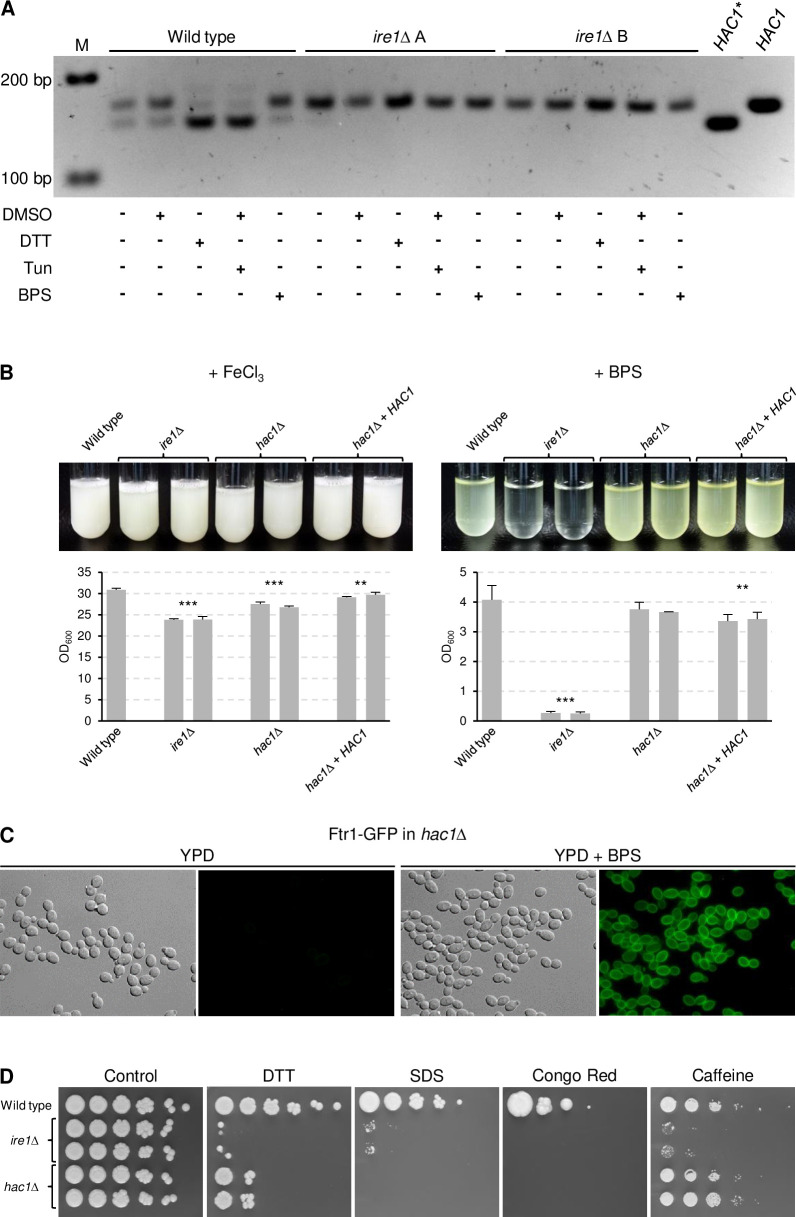
Fig 10. HAC1 is not required for growth of C. albicans under iron-limiting conditions.
(A) Detection of spliced and unspliced forms of HAC1 mRNA in the wild-type strain SC5314 and ire1Δ mutants. The strains were grown in the absence or presence of 5 mM DTT, 4 μg/ml tunicamycin, or 500 μM BPS as described in Materials and methods. Since tunicamycin is dissolved in DMSO, the cultures were also treated with a corresponding amount of DMSO alone as an additional control to no treatment. Total RNA was isolated from the cells and the region containing the HAC1 intron amplified by RT-PCR. The RT-PCR products were analyzed by agarose gel electrophoresis; the expected sizes are 162 bp and 143 bp, respectively, for the unspliced and spliced HAC1 transcripts. PCR products originating from plasmids containing wild-type HAC1 or the pre-spliced HAC1* are included for comparison in addition to size markers (M). (B) The wild-type strain SC5314, ire1Δ mutants, and hac1Δ mutants and complemented strains were grown in minimal medium with (+ FeCl3) or without iron (+ BPS). Cultures were photographed after 4 days of growth at 30°C (top panels) and the optical densities of three independent cultures of each strain determined (bottom panels showing means and standard deviations). Results for both independently generated series of strains are shown separately; data were combined for statistical analysis. Significant differences from the wild type are indicated by stars (***, p < 0.001; **, p < 0.01). (C) Overnight cultures of hac1Δ mutants containing a GFP-tagged FTR1 allele were inoculated in fresh YPD medium without or with BPS and grown for 5 h at 30°C. Cells were imaged by DIC and fluorescence microscopy. (D) Comparison of the sensitivities of ire1Δ and hac1Δ mutants to ER and cell membrane/wall stress. YPD overnight cultures of the strains were serially 10-fold diluted, spotted on YPD plates without (control) or with 20 mM DTT, 0.04% SDS, 50 μg/ml Congo Red, or 15 mM caffeine and incubated for 4 days at 30°C. Both independently generated series of mutants are shown in (A), (B), and (D). The two independently generated hac1Δ mutants containing Ftr1-GFP behaved identically and only one of them is shown in (C).