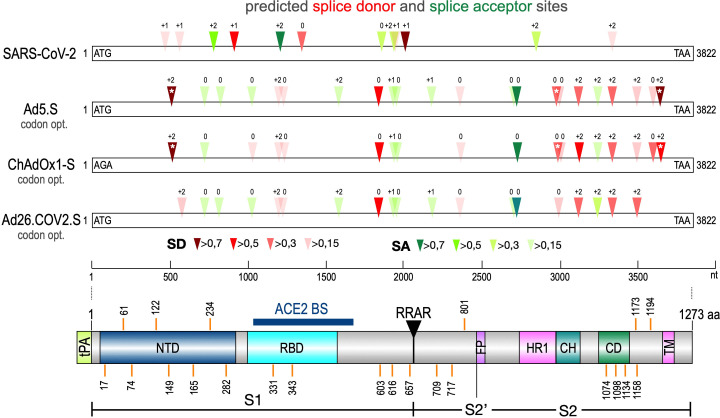
Figure 1. Splice site prediction within the Spike open reading frames.
Splice site prediction was carried out by using SpliceRover. Splice donor sites are given in red, splice acceptor sites in green. SpliceRover calculates splice sites with a score between 0 and 1, but only splice sites with >0.15 were displayed. Sites were displayed in four colors as indicated. Splice sites were numbered with ‘0’, ‘+1’, or ‘+2’, to indicate how the open reading frame is disrupted. Therefore, all splice events between splice sites with identical numbers will be in-frame, while all splice reactions between unequal numbers will result in out-of-frame fusions. Below, the protein domain structure of Spike is displayed to explain, which domains are being deleted by splice events (NTD: N-terminal domain, RBD: (ACE2) receptor-binding domain, RRAR: furin cleavage site, HR1: heptad repeat1, CH: central helix, CD: connector domain, TM: transmembrane domain). White asterisks mark three splice donor sites that are present in the codon-optimized Spike reading frame of the Vaxzevria but not of the Janssen vaccine.