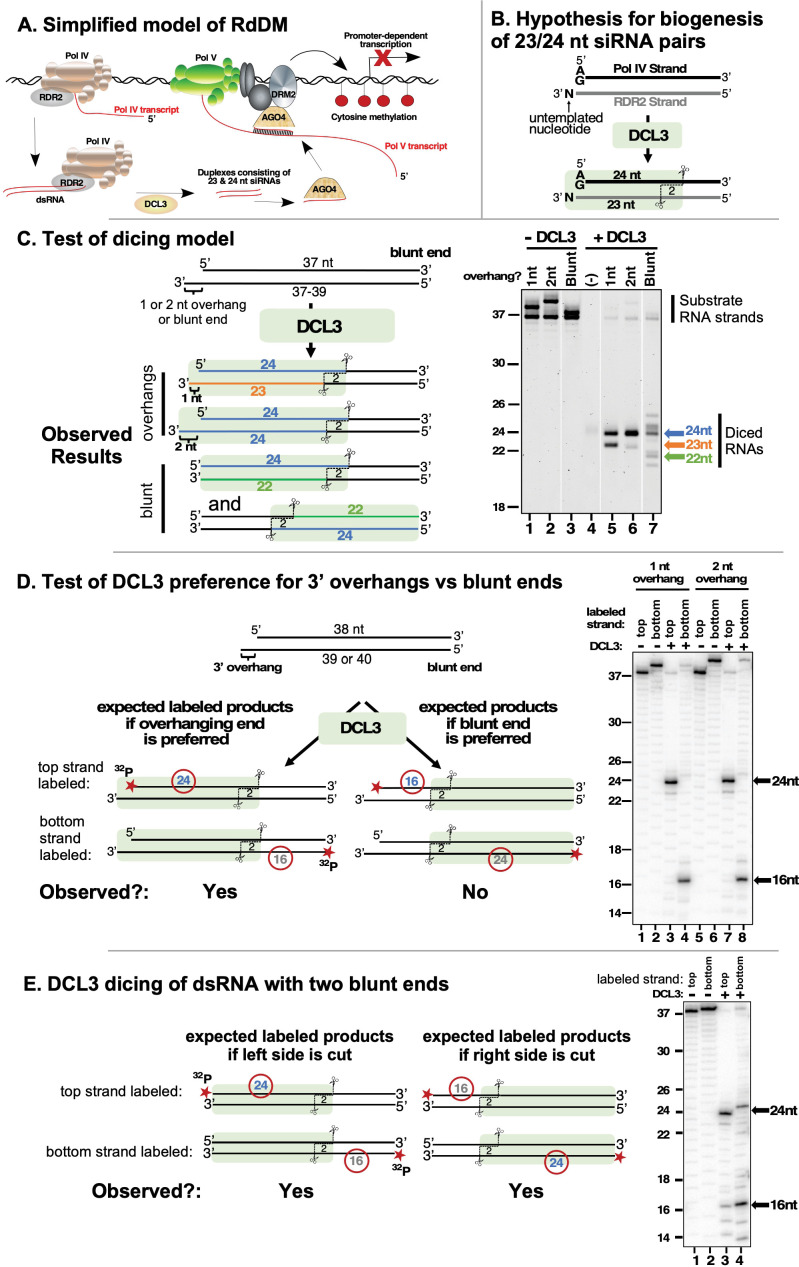
Figure 1. DCL3 preferentially dices double-stranded RNAs with 3′ overhangs.
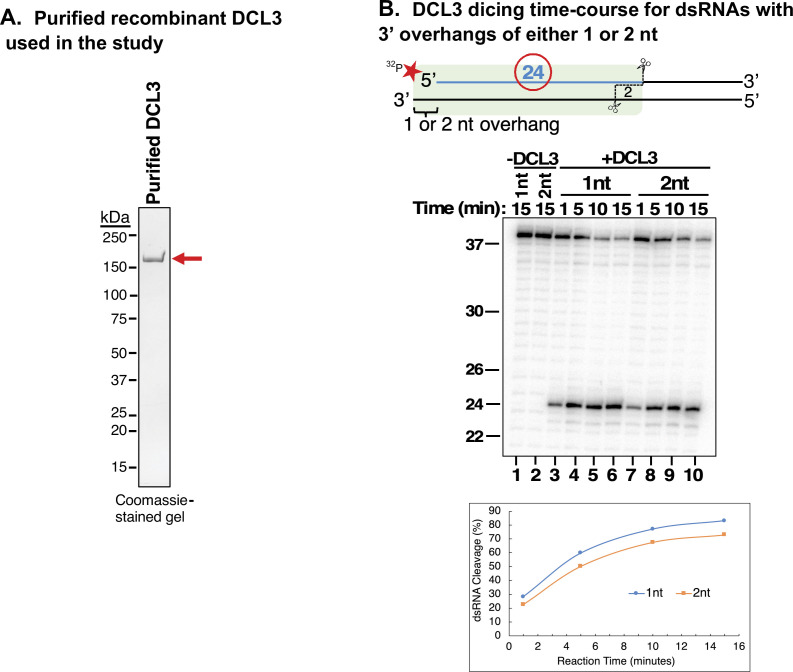
(A) A simplified model of RNA-directed DNA methylation (RdDM) highlighting the roles of Pol IV, Pol V, RDR2, DCL3, and AGO4. (B) Model depicting the hypothesis that DCL3 dicing of dsRNA precursors can yield a 24 nt siRNA from the 5′ end of the Pol IV transcript paired to a 23 nt siRNA from the RDR2 3′ end. Pol IV transcripts tend to begin with A or G and RDR2 transcripts often have an untemplated nucleotide (N) at their 3′ termini. Green shading depicts DCL3 and its interaction with the left side of the dsRNA precursor. (C) A test of the model shown in (B). A 37 nt top strand was annealed to 37, 38, or 39 nt bottom strand to form dsRNA substrates with two blunt ends or a left-side 1 or 2 nt 3′ overhang on the bottom strand (see Supplementary file 1 for RNA strand sequences). Resulting dsRNAs (50 nM) were then incubated with 25 nM of affinity purified recombinant DCL3 (see Figure 1—figure supplement 1 – (A)). RNAs were then resolved by denaturing polyacrylamide gel electrophoresis (PAGE) and visualized using SYBR Gold staining. Lane 4 is a control that includes DCL3 but no RNA. (D) DCL3 prefers 3′ overhangs. Dicing reactions were conducted as in (C), but with either the top strand (37 nt) or bottom strand (38 or 39 nt) 5′ end-labeled with 32P and the final concentration of dsRNAs being 25 nM. In each case, a non-radioactive monophosphate is also present at the 5′ end of the complementary strand. Following incubation with (lanes 3, 4, 7, and 8) or without (lanes 1, 2, 5, and 6) DCL3, RNAs were resolved by denaturing PAGE and visualized by phosphorimaging. A related experiment comparing time courses of DCL3 cleavage for substrates with 1 or 2 nt overhangs is shown in Figure 1—figure supplement 1 – (B). (E) DCL3 cuts from both ends of precursors that have two blunt ends. Dicing reactions of 5′ end-labeled dsRNAs were conducted as in (E) but with precursors that lack a 3′ overhang at one end.