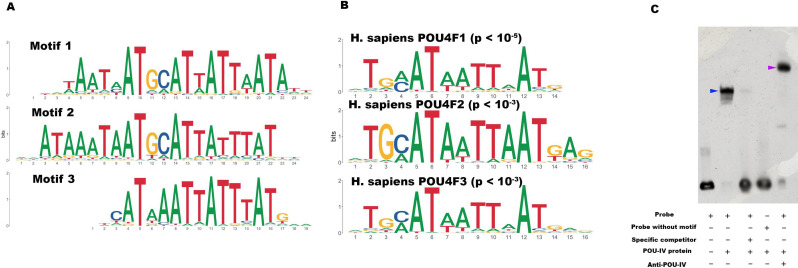
Figure 7. POU-IV-binding motifs are conserved across Cnidaria and Bilateria.
(A) Motifs enriched in Nematostella vectensis POU-IV chromatin immunoprecipitation sequencing (ChIP-seq) peaks. (B) Homo sapiens POU motifs resulting from sequence alignment and comparison against the Jaspar database. The p-value reported corresponds to the highest p-value for any of the three N. vectensis POU4 motifs found. (C) Electrophoretic mobility shift assay (EMSA) using purified N. vectensis POU-IV protein and a 50 bp DNA probe containing the conserved core motif CATTATTAAT. Note that retardation of probe migration occurs in the presence of POU-IV protein (blue arrowhead; lane 2), indicative of formation of the protein-DNA complex. Retardation is inhibited in the presence of an unlabeled competitor probe (‘specific competitor’; lane 3). Removal of the motif sequence in the probe (‘probe without motif’) abolishes retardation of probe migration by POU-IV (lane 4), demonstrating that the motif is necessary for formation of the protein-DNA complex. The mobility of the probe is further decreased in the presence of the anti-POU-IV antibody (purple arrowhead; lane 5), confirming that the protein bound to the probe is POU-IV.