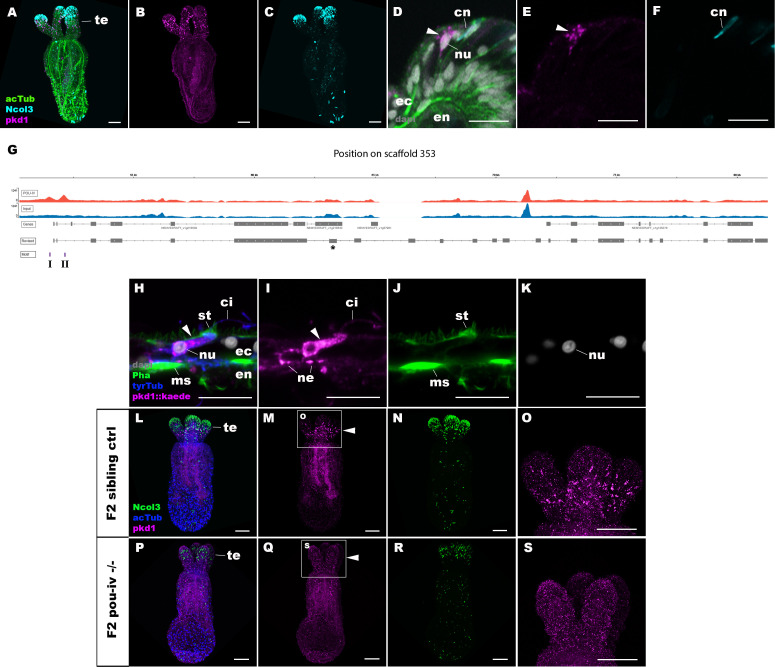
Figure 8. POU-IV activates the expression of polycystin 1 specifically in hair cells.
(A-F) Confocal sections of primary polyps labeled with an antisense riboprobe against polycystin 1 transcript (‘pkd1’) and antibodies against acetylated ∂-tubulin (‘acTub’) and minicollagen 3 (‘Ncol3”; Zenkert et al., 2011). Nuclei are labeled with DAPI (‘dapi’). A–C are side views of the animal with the oral opening facing up. Expression of polycystin 1 occurs exclusively in the ectoderm of the oral tentacles (te). (D–F) Side views of a polycystin 1-expressing epithelial cell (arrowhead) in the tentacular ectoderm (ec) with its apical surface facing up. Note that the cell body is localized apically and lacks minicollagen 3 expression. (G) A schematic of the polycystin 1 locus, showing the distribution of POU-IV ChIP DNA (‘POU-IV’) and input DNA from adult polyps. JGI gene models (‘Genes’) and the revised gene model based on RTPCR (‘Revised’) and the locations of the consensus POU-IV-binding motif – AT(A/T)ATT(A/T)AT – are numbered as I and II. X-axis shows the position along the genomic scaffold, and Y-axis shows the number of reads. * shows an exon whose sequence is missing in the publicly available Nematostella vectensis genome (v1.0; Putnam et al., 2007). (H–K) Confocal sections of an oral tentacle of a primary polyp injected with polycystin 1::kaede construct, labeled with an antibody against Kaede (‘pkd1::kaede’). Filamentous actin is labeled with phalloidin (Pha). The apical surface of the tentacular ectodermal epithelium is to the top. Note that the Kaede-positive cell (arrowhead) has an apical cilium (ci) and stereovilli (st), a central nucleus (nu), and basal neurites (ne), exhibiting morphological hallmarks of a hair cell. No other cell types were found to be Kaede-positive. L–S: Confocal sections of a homozygous pou-iv mutant (‘F2 pou-iv -/-’, P-S) and its sibling control (F2 pou-iv +/+ or pou-iv +/-, ‘F2 sibling ctrl’, L–O) at the primary polyp stage, labeled with an antisense riboprobe against polycystin 1 transcript (‘pkd1’) and antibodies against acetylated ∂-tubulin (‘acTub’) and minicollagen 3 (‘Ncol3’; Zenkert et al., 2011). Panels show side views of the animal with the oral opening facing up. Animals lacking mature cnidocysts based on Ncol3 staining were assumed to be pou-iv -/- mutants; animals with mature cnidocysts were assumed to be pou-iv +/+ or pou-iv +/-. O and S are magnified views of tentacles boxed in M and Q, respectively. Note that cell-type-specific expression of polycystin 1 in tentacular ectoderm (arrowhead in M; O) is absent in the POU-IV null mutant (arrowhead in Q; S), demonstrating that POU-IV is necessary for polycystin 1 expression. Abbreviations: en, endoderm; cn, cnidocyst; nu, nucleus; ms, muscle fiber. Scale bar: 50 µm (A–C, L–S); 10 µm (D–F, H–K).