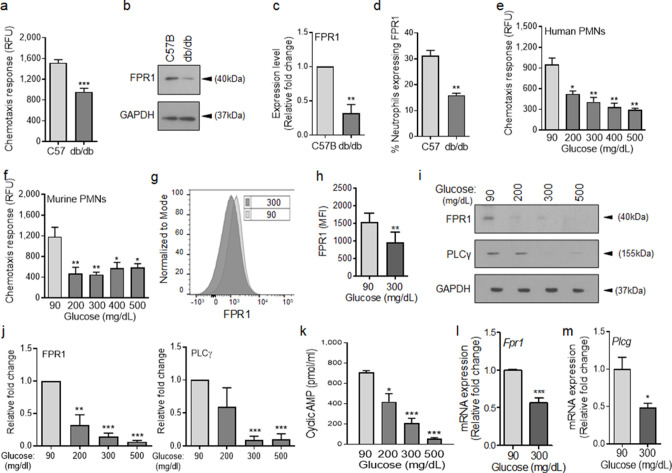
Figure 2. Chemotactic response is impaired in diabetic neutrophils through FPR.
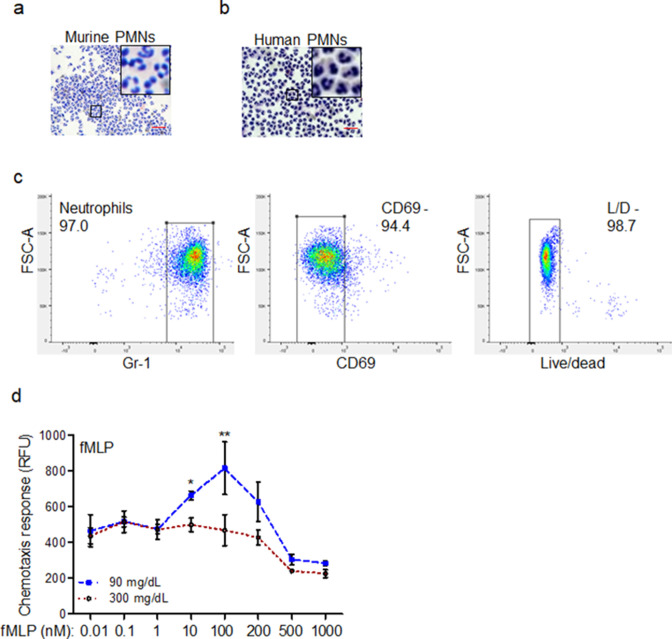
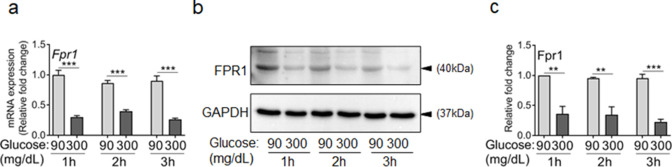
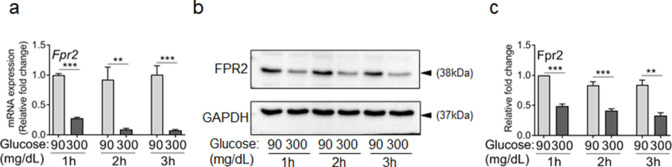
(a–b) Neutrophils were isolated from the peripheral blood of C57BL/6 and db/db animals to assess: (a) their ability to chemotax toward 100 nM fMLP, or (b) for the expression of FPR1 by Western blotting. (c) Densitometry values associated with (b) are plotted as Mean ± SEM (N = 4 blood pools/group, each blood pool was from 4 mice). (d) Equal number of neutrophils (isolated from Day 1 C57B and db/db wounds) were assessed for the surface expression of FPR1 on neutrophils by flow cytometry (N = 3 mice/group). (e–f) Purified neutrophils from peripheral blood of non-diabetic individuals (e), or C57BL/6 bone marrow (f), were exposed to media containing glucose in normal range (90 mg/dl) or in diabetic range (200–500 mg/dl) for 1 hr to assess their ability to chemotax toward 100 nM fMLP. Data are plotted as Mean ± SEM. (N > 4). (g–h) Neutrophils from C57BL/6 bone marrow were exposed to glucose in normal range (90 mg/dl) or in diabetic range (300 mg/dl) for 1 hr and assessed for surface expression of FPR1 by flow cytometry. A representative histogram is shown in (g) and the corresponding tabulated data, plotted as Mean ± SEM is shown in (h) (N = 3). (i–j) Murine neutrophils (from C57B bone marrow) were exposed to glucose in normal or diabetic range (90 mg/dl or 300 mg/dl) for 1 hr and assessed for the expression of indicated proteins by Western blotting. Representative Western blots are shown in (i) and corresponding densitometry values, plotted as Mean ± SEM, are shown in (j). (N ≥ 3 independent experiments). (k–m) Murine neutrophils exposed to normal or diabetic glucose, as described for (g–h), were assessed for Cyclic AMP production by ELISA (k), and for mRNA of Fpr1 and Plcγ by RT-PCR (l-m). (N ≥ 3, ns = not significant, *p < 0.05, **p < 0.01, ***p < 0.001. Statistical analyses between groups were conducted by One-way ANOVA with additional post hoc testing, and pair-wise comparisons between groups were performed or by unpaired Student’s t-test).