Figure 5.
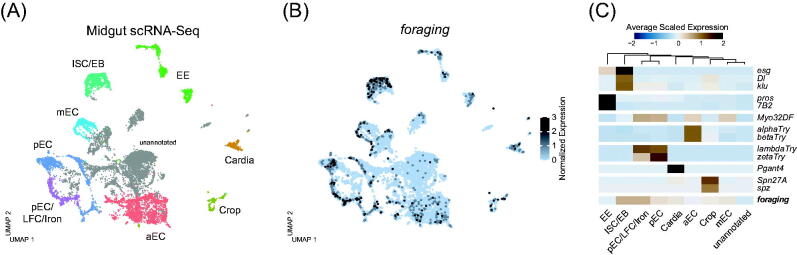
(A) UMAP plot of single-cell RNA sequencing from the adult midgut (data from Hung et al., 2020). Each point represents the transcriptome of a single cell. Cells are clustered based on similarity of gene expression. Distinct cell types are represented by different colors. EE: enteroendocrine cells; ISC: intestinal stem cells; EB: enteroblasts; LFC: large flat cells; Iron: iron cells; aEC: anterior enterocytes; mEC: middle enterocytes; pEC: posterion enterocytes. (B) Expression of foraging in the single-cell midgut Atlas. Cells are color coded according to the level of normalized expression. (C) Heatmap showing the average scaled expression of cell type marker genes across each cell cluster. Initialisms are defined in A. foraging is most enriched in the ISC/EB and pEC/LFC/Iron clusters. EE cells are marked by pros and 7B2. ISC/EB are marked by esg, Dl, and klu. All ECs are marked by Myo32DF, and the anterior to posterior access is delineated by a series of trypsin coding genes (alphaTry, betaTry, lambdaTry, zetaTry, among others). The cardia is marked by Pgant4, and the crop by Spn27A and spz. [Please refer to the online version for colors.]