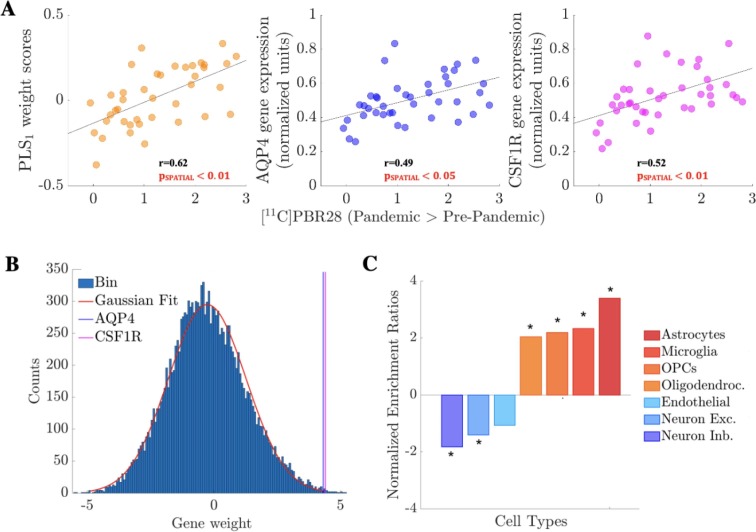
Fig. 4.
Imaging transcriptomics analyses. (A) First component of the PLS analyses (PLS1; weight scores), AQP4 (astrocyte marker) and CSF1R (microglia marker) gene expression (normalized units) plotted against [11C]PBR28 contrast t-stat (Pandemic > Pre-pandemic; see Fig. 1) in the 41 regions of left hemisphere regions (34 cortical plus 7 subcortical regions) of the Desikan-Killiany atlas. Genes with positive weights in PLS1 have higher-than-average expression where [11C]PBR28 showed the largest increases and lower-than-average expression in regions with minimal changes. Of note, pSPATIAL is obtained via spatial permutation testing (spin test) to account for the inherent spatial autocorrelation of the imaging data (see Supplementary materials). (B) Histogram (150 bins) of gene weights in PLS1 with the highly-ranked positions of AQP4 and CSF1R highlighted (scores 4.31 and 4.40 respectively). (C) Brain cell-type gene set enrichment analysis. Positive normalized enrichment ratios (in orange-red shades) indicate enrichment for genes of a certain cell-type among those genes with high expression in regions with the largest increases in [11C]PBR28 signal (positive weights in PLS1). Negative normalized enrichment ratios (in blue shades) indicate enrichment for genes of a certain cell-type among genes with high expression in regions with minimal or negligible increases in [11C]PBR28 signal (negative weights in PLS1). *=p < 0.05, after FDR correction for the total number of cell-types tested. Abbreviations: OPCs = Oligodendrocyte Precursor Cells; Oligodendroc. = Oligodendrocytes; Neuron Exc. = Excitatory Neurons; Neuron Inb. = Inhibitory Neurons.