Fig 2. Flexibility of PEDAR in programming larger deletion and insertion in HEK293T cells.
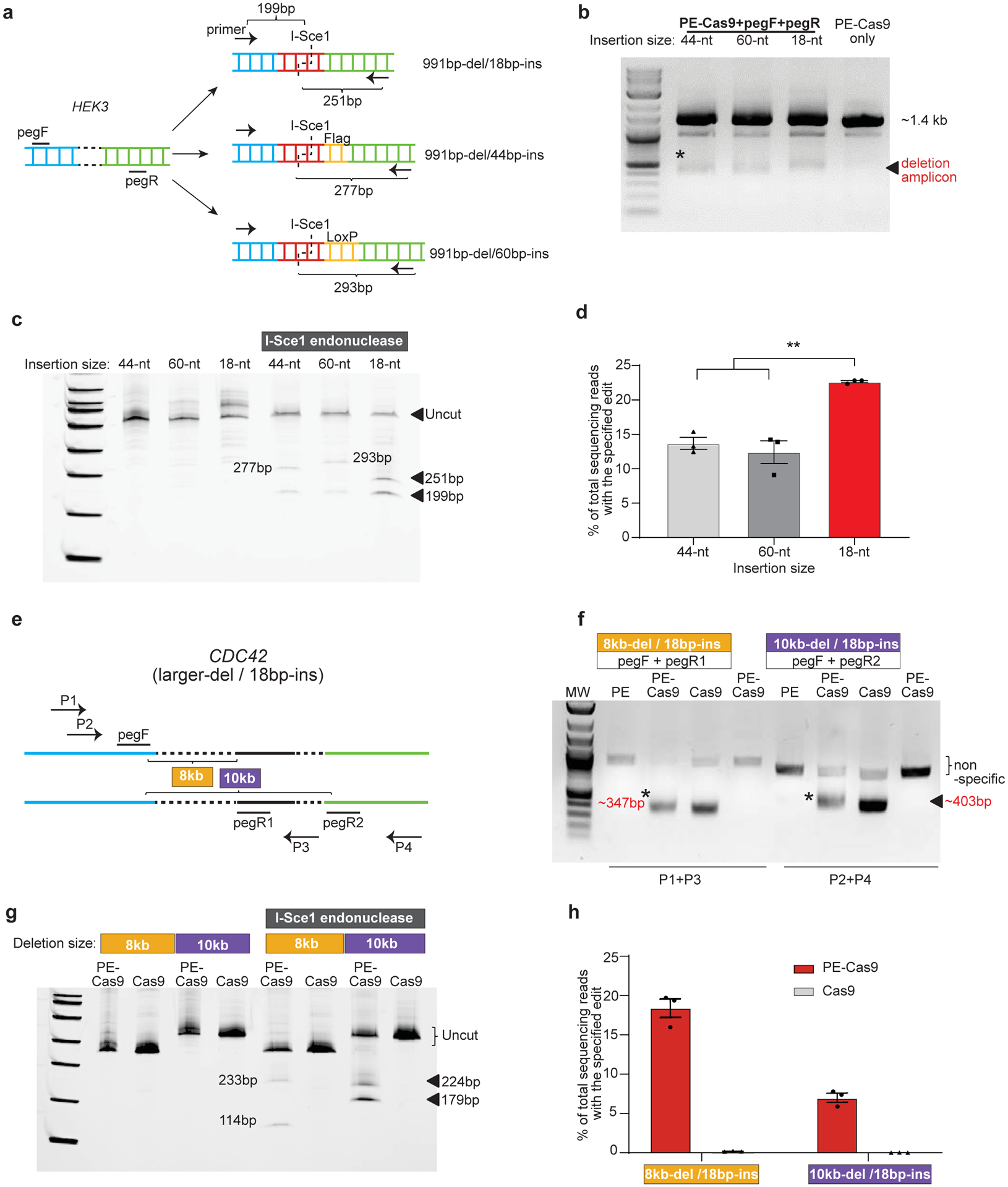
a, Insert DNA fragments of variable lengths (18-bp, 44-bp, and 60-bp) to the target site at HEK3 locus. pegRNAs and primers for amplifying the target site are as shown. The expected sizes of digestion products after I-Sce1 treatment are shown. b, Amplification of target genomic region using primers spanning the cut sites at HEK3 locus. The deletion amplicons are denoted with *. Cells transfected with PE-Cas9 alone serves as negative control. c, Deletion amplicons from groups shown in (b) were incubated with or without I-SceI endonuclease and analyzed in 4–20% TBE gel. Digested products are marked by arrows with expected sizes. The original amplicon is marked as “uncut”. d, Deep sequencing of deletion amplicons shown in (b). Bar chart shows accurate deletion-insertion rate. Data represent mean ± SEM (n = 3 biologically independent samples). P=0.0024 (**), two-tailed t-test. e, Test the efficiency of PEDAR in mediating larger deletions. Paired pegRNAs spaced ~8-kb (pegF+pegR1) or 10-kb (pegF+pegR2) apart were designed as indicated to target the CDC42 locus. Primers used to amplify the target genomic regions are as marked (P1+P3 and P2+P4). f, Target genomic region was amplified using the primers indicated in (e). Cells transfected with PE-Cas9 alone serve as negative control. The deletion amplicons are marked with expected sizes (denoted with *). g, Deletion amplicons from Cas9- or PE-Cas9-treated groups shown in (f) were incubated with or without I-SceI endonuclease and analyzed in 4–20% TBE gel. Digested products are marked with expected sizes. The original amplicon is marked as “uncut”. h, Deep sequencing of deletion amplicons shown in (f). Bar chart shows rate of accurate deletion-insertion events. Data represent mean ± SEM (n = 3 biologically independent samples).
