Fig 3. PEDAR generates in-frame deletion to restore mCherry expression in TLR reporter cells.
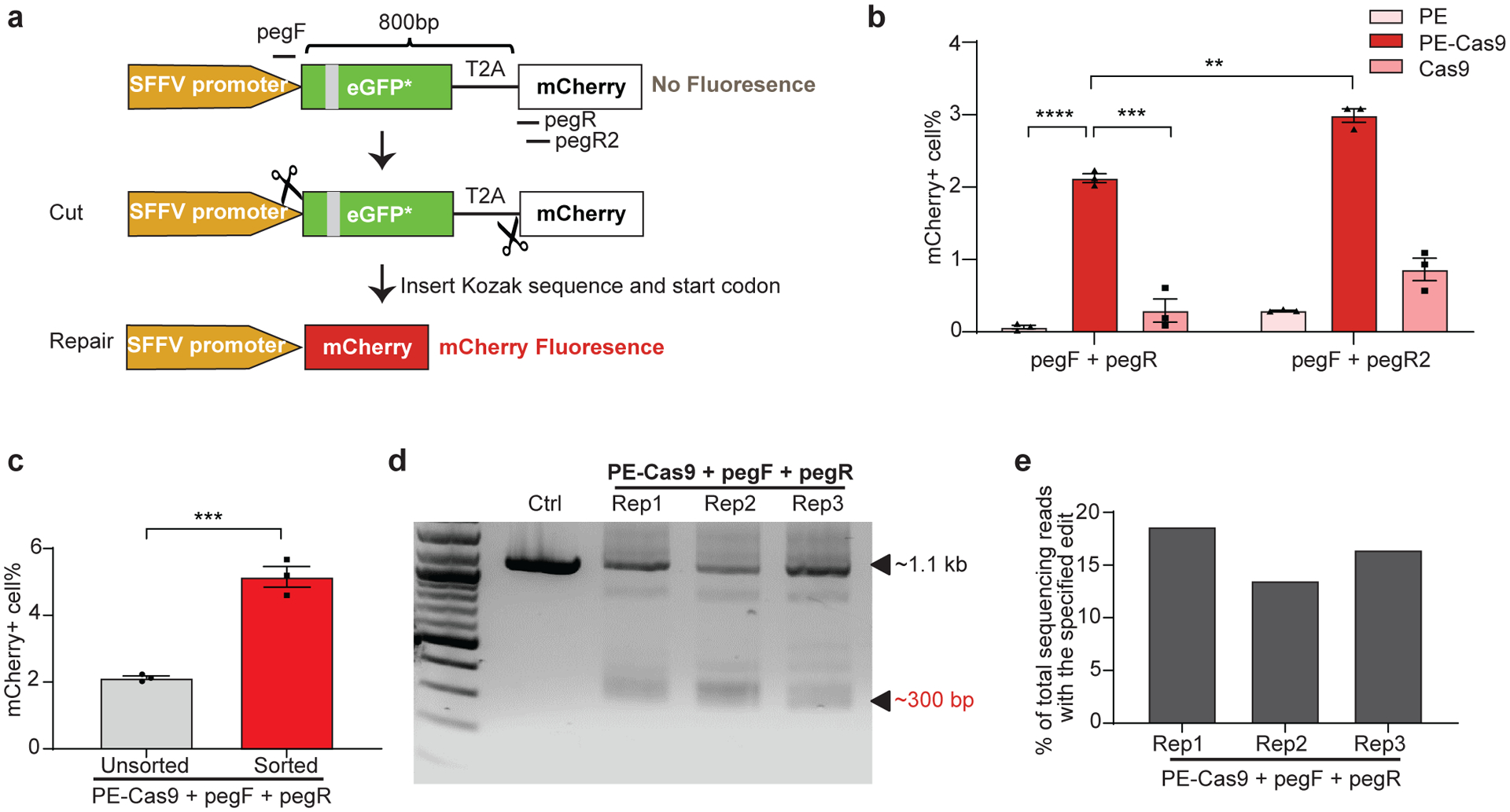
a, Diagram of the TLR reporter system. GFP sequence is disrupted by an insertion (grey). Deleting the disrupted GFP sequence and inserting Kozak sequence and start codon will restore mCherry protein expression. b, TLR reporter cells transfected with indicated paired pegRNAs along with PE, PE-Cas9, or Cas9 were analyzed by flow cytometry, and the percentage of mCherry positive cells are shown among different groups. Data represent mean ± SEM (n = 3 biologically independent samples). P=0.00000636 (****), =0.0004 (***), =0.0015 (**), two-tailed t-test. c, mCherry positive cell rate before and after sorting of cells with high transfection level. A plasmid expressing GFP was co-transfected with paired pegRNAs and PE-Cas9 into TLR cells. Three days later, cells with high GFP expression were selected for analyzing mCherry signal by flow cytometry. Data represent mean ± SEM (n = 3 biologically independent samples). P=0.0007 (***), two-tailed t-test. d, TLR reporter cells edited by PEDAR were selected by flow cytometry (for mCherry signal) and subjected to PCR amplification using primers spanning the two cut sites. The amplicon with the desired deletion is ~300 bp compared to a ~1.1-kb PCR products in control group. Rep: replicate; Ctrl: untreated TLR reporter cells. e, Efficiency of accurate deletion-insertion in three PEDAR-edited replicates (Rep 1–3) measured by deep sequencing of the deletion amplicons shown in (d).
