Fig 4. PEDAR corrects the pathogenic insertion in a Tyrosinemia I mouse model.
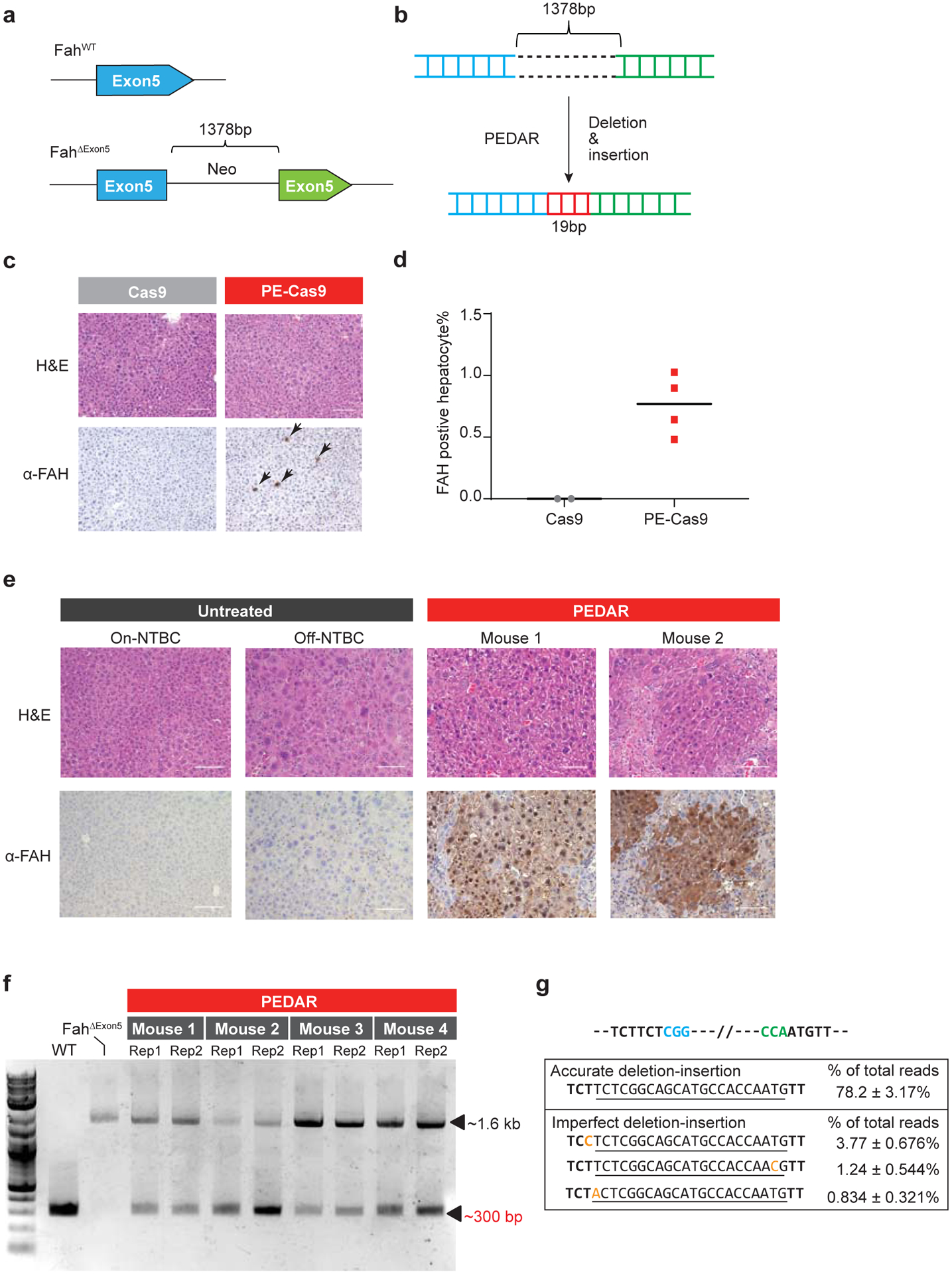
a, The Tyrosinemia I mouse model, referred to as FahΔExon5, was derived by integrating a ~1.38-kb neo expression cassette at exon 5 of the Fah gene. b, Diagram showing the application of PEDAR to delete the ~1.38-kb insertion and concurrently repair the target region by inserting a 19-bp DNA fragment (marked in red). c, Immunohistochemistry staining and Hematoxylin and Eosin staining (H&E) of mouse liver sections seven days after injection of dual pegRNAs with Cas9 or PE-Cas9. FAH positive hepatocytes are pointed by arrows. Scale bar=100μm. d, Quantification of FAH expressing hepatocytes shown in (c). n=2 (Cas9-treated group), n=4 (PE-Cas9-treated group). e, Immunohistochemistry and H&E staining of mouse liver sections 40 days after injection of PE-Cas9 with dual pegRNAs. Mice (n=4) were kept off NTBC. Mouse 1 and 2 denote two representative mice from the treatment group. The liver sections from untreated Fah ΔExon5 kept on or off NTBC serve as negative controls. Scale bar=100μm. f, Amplification of exon 5 of Fah gene from mouse livers 40 days post injection of PE-Cas9 and paired pegRNAs. The corrected amplicon size is around ~300 bp, compared to a ~1.6-kb amplicon without deletion. Four mice in treated group and two liver lobes (denoted as Rep 1 and 2) per mouse were analyzed. WT: wild type C57BL/6J mouse. Fah ΔExon5: untreated Fah ΔExon5 mouse. g, Accurate correction rate and the top-three imperfect editing events identified by deep sequencing. Two PAM sequences are in blue and green. The 22-bp intended insertion (19-bp deletion fragment plus a 3-bp unintentionally deleted sequence) is underlined. Mutated nucleotides in imperfect editing sequences are highlighted in yellow. Data represent mean ± SD (n = 8, two liver lobes/mouse; four mice in total).
