Figure 2. Concurrent programming of deletion and insertion using PRIME-Del.
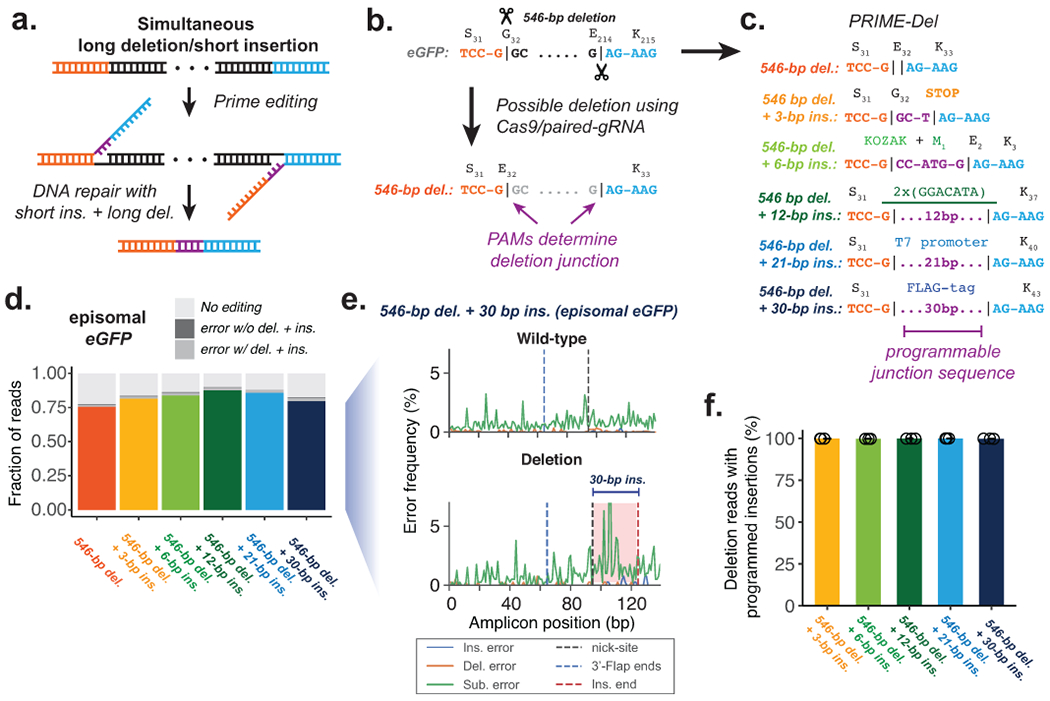
a. Schematic of strategy, with reverse complementary sequences corresponding to the intended insertion in purple. b. Conventional strategy for deletion with Cas9 and pairs of gRNAs. Potential deletion junctions are restricted by the natural distribution of PAM sites. c. Pairs of pegRNAs were designed to encode five insertions, ranging in size from 3 to 30 bp, together with a 546 bp deletion in eGFP. d. Estimated deletion efficiencies and indel error frequencies (with or without intended deletion) in using these pegRNA pairs to induce concurrent deletion and insertion in HEK293T cells (averaged over replicates; n = 3). e. Representative insertion, deletion and substitution error frequencies plotted across sequencing reads from concurrent 546-bp deletion and 30-bp insertion condition. Plots are from single-end reads without UMI correction. Note that only one of the two 3’-DNA-flaps is covered by the sequencing read in amplicons lacking the deletion (labeled as ‘wild-type’). f. The percentage of reads containing the programmed deletion that also contain the programmed insertion. Error bars represent standard deviation for at least three replicates.
