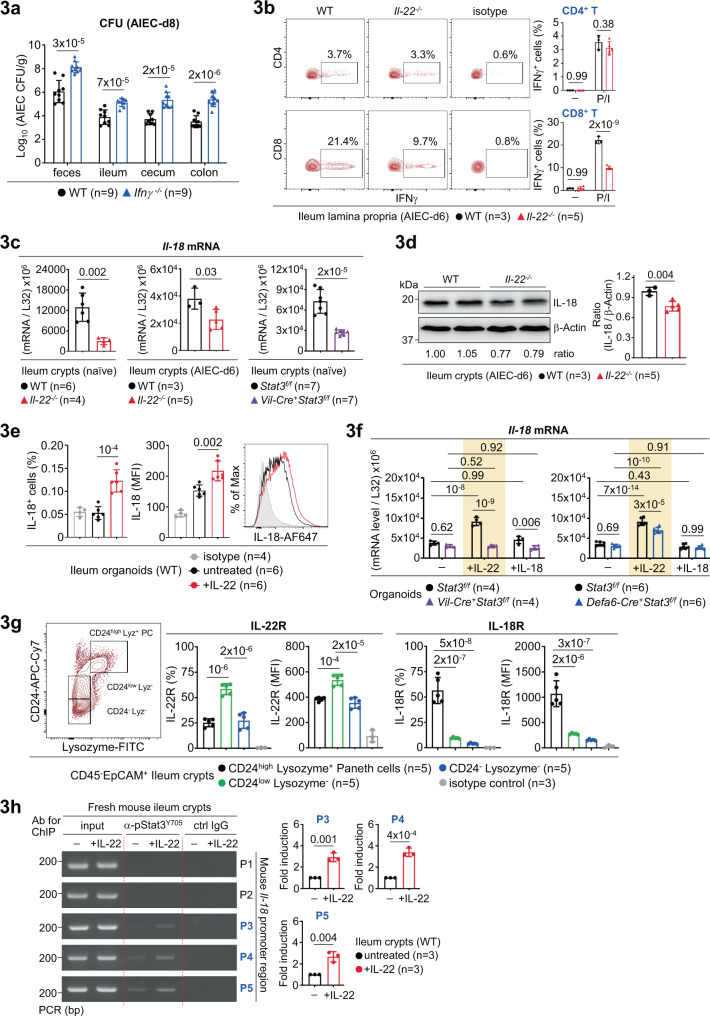
Fig. 3. IL-22 transcriptionally induces epithelial IL-18 via phospho-Stat3Y705.
a Colony-forming unit (CFU) of AIEC was analyzed in feces and intestinal tissues isolated from the indicated mice at day-8 (d8) post infection. b Flow cytometry analysis of PMA/Ionomycin (P/I)-stimulated ileum lamina propria cells for intracellular IFNγ in CD4+ or CD8+ T cells in AIEC-infected mice. c Quantitative real-time PCR analysis of ileum crypts from the indicated mice for Il-18 mRNA levels. d Western blot analysis of ileum crypts from the indicated mice for IL-18. Quantification and the ratio (IL-18 to β-actin) of protein bands are indicated. e Flow cytometry analysis of IL-22-stimulated ileum organoids for intracellular IL-18. f Quantitative real-time PCR analysis of IL-22 or IL-18-stimulated ileum organoids, derived from the indicated mice, for Il-18 mRNA levels. g Flow cytometry analysis of naïve wild-type ileum crypts for IL-22R or IL-18R expression levels in the indicated epithelial subsets. h Chromatin immunoprecipitation (ChIP) and PCR analyses of IL-22-stimulated ileum crypts for the binding of phospho-Stat3Y705 to the mouse Il-18 promoter regions (P1~P5). Fold induction is calculated based on PCR signal strength of pStat3-bound Il-18 promoter region (P3,P4,P5) before and after IL-22 stimulation. Each symbol in bar graphs represents an ileum crypt sample (c, d, g, h), tissue sample (a, b), or organoid culture (e, f), derived from one mouse. Data shown are representative (b, d, g, h) or combined (a, c, e, f) results from two independent reproducible experiments. Statistical significance is indicated using unpaired two-tailed t test (a, c, d, e, h), One-way ANOVA with Sidak’s multiple comparisons test (g), or Two-way ANOVA with Tukey’s multiple comparisons test (b, f). Data are presented as mean ± SD. Source data are provided as a Source Data file.