Summary
Tumor purity is the percentage of cancer cells within a tissue section. Pathologists estimate tumor purity to select samples for genomic analysis by manually reading hematoxylin-eosin (H&E)-stained slides, which is tedious, time consuming, and prone to inter-observer variability. Besides, pathologists' estimates do not correlate well with genomic tumor purity values, which are inferred from genomic data and accepted as accurate for downstream analysis. We developed a deep multiple instance learning model predicting tumor purity from H&E-stained digital histopathology slides. Our model successfully predicted tumor purity in eight The Cancer Genome Atlas (TCGA) cohorts and a local Singapore cohort. The predictions were highly consistent with genomic tumor purity values. Thus, our model can be utilized to select samples for genomic analysis, which will help reduce pathologists' workload and decrease inter-observer variability. Furthermore, our model provided tumor purity maps showing the spatial variation within sections. They can help better understand the tumor microenvironment.
Keywords: tumor purity, genomic sequencing, tumor microenvironment, spatial omics, digital histopathology, whole-slide images, multiple instance learning, deep learning, digital pathology, computational pathology
Highlights
-
•
MIL model successfully predicts a sample's tumor purity from histopathology slides
-
•
MIL model learns to spatially resolve tumor purity from sample-level labels
-
•
Tumor purity varies spatially within a sample
-
•
Pathologists’ region selection is vital for correct percentage tumor nuclei estimation
The bigger picture
Given some big data and coarse-level labels, extracting fine-level information is a demanding yet rewarding challenge in data science. This study develops a machine learning model utilizing big data and exploiting coarse-level labels to reveal fine-level details within the data. Although it can be applied to different data science tasks with enormous data and coarse labels, we applied it to a computational histopathology task with gigapixel histopathology slides and sample-level labels. Specifically, the model revealed spatial resolution of tumor purity within histopathology slides using only sample-level genomic tumor purity values during training. This can also be extended to other omics features, providing precious information about cancer biology and promising personalized, precision medicine. Such studies are of great clinical importance in discovering imaging biomarkers and better understanding the tumor microenvironment.
Selecting a sample with sufficient tumor content is crucial for the proper operation of sequencing methods. This study developed a deep learning model predicting the percentage of cancer cells (tumor purity) within a tissue section from its digital histopathology slides to support pathologists in sample selection for genomic sequencing. The model successfully predicted tumor purity in eight different cancers and produced tumor purity maps showing the spatial variation within sections without requiring pixel-level annotations from pathologists during training.
Introduction
High-throughput genomic analysis has become an indispensable tool for cancer research and has enabled precision oncology.1,2 One of the crucial factors affecting the quality of genomic analysis is the proportion of cancer cells in the samples.3 Tumors consist of a complex mixture of cells, such as cancer cells, normal epithelial cells, stromal cells, and infiltrating immune cells.4 The proportion of cancer cells in a section can significantly influence the accuracy of not only sequencing experiments but also precision oncology. The subjective estimates of the percentage of cancer cells within a tissue section—or tumor purities—are routinely evaluated by pathologists.5
The tumor purity affects both high-throughput data acquisition and analysis. To detect genetic variations of a tumor sample by next-generation sequencing, the sample needs to have sufficient cancer cells.6, 7, 8 Therefore, an accurate tumor purity estimation is of great clinical importance. A sample with low tumor purity, for example, may lead to a false-negative test result, potentially resulting in missed therapeutic opportunities.6 Besides, the genomic analysis should incorporate the tumor purity to account for normal cell contamination, which can have confounding effects on analysis results.5,9, 10, 11, 12, 13, 14 A novel immunotherapy gene signature missed by traditional methods, for example, was discovered using a differential expression analysis incorporating tumor purity.5 The tumor purity is also associated with clinical variables.15, 16, 17 Low tumor purity, for instance, was associated with poor prognosis in glioma,15 colon cancer,16 and gastric cancer.17 Moreover, tumor purity was a promising predictor for therapeutic response in colon cancer16 and gastric cancer.17
A pathologist estimates tumor purity by reading hematoxylin and eosin (H&E)-stained histopathology slides. Essentially, the pathologist counts the percentage of tumor nuclei over a region of interest (ROI) in the slide. The tumor purity estimated in this way is referred to as percentage tumor nuclei in this study. The percentage tumor nuclei estimates are usually used for sample selection and interpretation of results in the molecular analysis. The pathologist can read any H&E-stained slide and estimate percentage tumor nuclei based on a cellular-level analysis. Thus, this approach is widely applicable, and it has a cellular-level resolution. However, counting tumor nuclei is tedious and time consuming. More importantly, there exists inter-observer variability between pathologists' estimates.6,18
Tumor purity can also be inferred from different types of genomic data, such as somatic copy number19, 20, 21, 22, 23, 24, 25 and mutations,26, 27, 28, 29, 30, 31 gene expression data,32, 33, 34, 35 and DNA methylation data.36, 37, 38, 39 The tumor purity obtained from these methods will be referred to as genomic tumor purity in this study. Genomic tumor purity values are usually used in genomics analysis to mitigate confounding effects of normal cell contamination40, 41, 42 and in correlational studies to investigate the associations between tumor purity and clinical variables.43 Nowadays, genomic tumor purity is accepted as “accurate” for downstream analysis.19,26,28,32,35 Genomic methods generally produce consistent values on different cancer datasets in The Cancer Genome Atlas (TCGA).5 However, they do not work well for the low-tumor-content samples. Furthermore, genomic methods cannot provide information on the spatial organization of the tumor microenvironment. Hence, both genomics methods and pathologists' slide reading approach have different strengths and limitations.
Pathologists routinely estimate percentage tumor nuclei in tissue sections. However, besides previously stated challenges, pathologists' estimates do not correlate well with genomic tumor purity values.5,13 To assist pathologists, this study develops a machine learning model that predicts the tumor purity from H&E-stained histopathology slides such that the predictions are consistent with the genomic tumor purity values. In addition to giving accurate tumor purity measurements, our model is cost-effective compared with genomics methods. It also provides information about the spatial organization of the tumor microenvironment.
Two types of machine learning models can be utilized to predict tumor purity from digital histopathology slides: patch-based models and multiple instance learning (MIL) models. The patch-based models require pathologists' pixel-level annotations showing whether each pixel is cancerous or normal. Although different studies employed this approach for tumor purity prediction,44, 45, 46, 47, 48, 49 they had limited coverage since pixel-level annotations are rarely available, expensive, and tedious. On the other hand, the MIL models do not require pixel-level annotations. Instead, they use sample-level labels, which are weak labels providing only aggregate information rather than pixel-level information. However, they can easily be collected from pathology reports, electronic health records, or different data modalities. The MIL models were successfully used in various digital pathology tasks,50, 51, 52 whereas this is the first study using the MIL approach to predict tumor purity. This study uses sample-level genomic tumor purity values as labels during training and does not require tedious pixel-level annotations by pathologists.
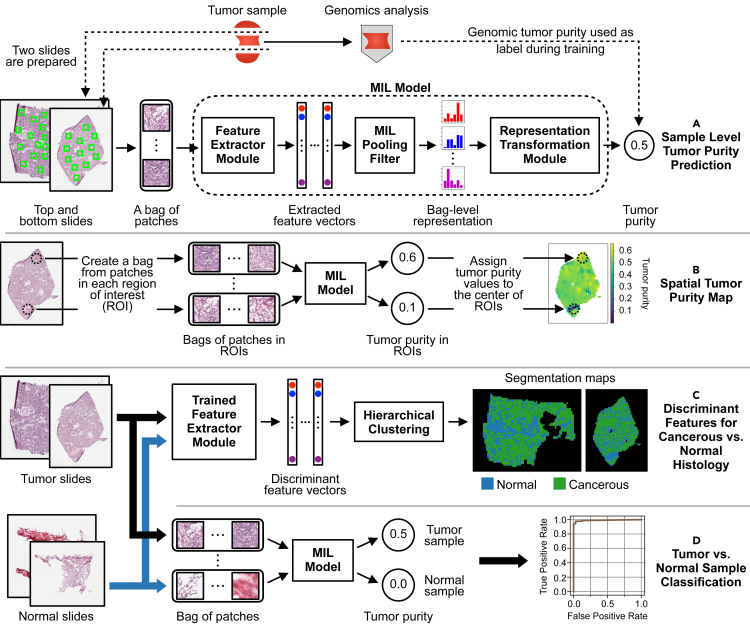
We formulate predicting tumor purity of a sample from its H&E-stained histopathology slides as an MIL task (Figure 1A). The sample's top and bottom slides are cropped into many patches, and these patches are collected to form a bag. Then, the task is to predict the bag-level label of tumor purity. To achieve this task, we developed a novel MIL model with a “distribution” pooling filter (see experimental procedures for details).
Figure 1.
A novel MIL model predicts sample-level tumor purity from H&E-stained digital histopathology slides
(A) Our model accepts a bag of patches randomly cropped from the top and bottom slides of a sample as input and predicts the sample's tumor purity at its output. The feature extractor module extracts a feature vector for each patch inside the bag. The MIL pooling filter, namely distribution pooling, summarizes extracted features into a bag-level representation by estimating marginal feature distributions. Finally, the bag-level representation transformation module predicts the sample-level tumor purity. We use tumor purity values inferred from genomic sequencing data by ABSOLUTE19 as ground-truth labels during training.
(B) We obtain a spatial tumor purity map for a slide by inferring tumor purity over each 1-mm2 ROI within the slide in a sliding window fashion. The map shows the variation of tumor purity over the slide.
(C) Our MIL model learned discriminant features for cancerous versus normal histology from sample-level genomic tumor purity labels without requiring exhaustive annotations from pathologists. We used discriminant features to obtain cancerous versus normal segmentation maps for tumor slides. Trained feature extractor module extracts features of patches from tumor and normal slides of a patient. Then, segmentation maps are obtained by hierarchical clustering over the extracted feature vectors.
(D) Our MIL model successfully classifies samples into tumor versus normal.
Our MIL models successfully predicted sample-level tumor purity in different TCGA cohorts and a local Singapore cohort. The predictions were consistent with genomic tumor purity values (Figure 2). Besides, we obtained spatially resolved tumor purity maps showing the variation of tumor purity over the slides (Figures 1B and 4). We also showed that our MIL models learned discriminant features for cancerous versus normal histology (Figures 1C and 5) and classified samples into tumor versus normal almost perfectly in all cohorts (Figures 1D and 3B).
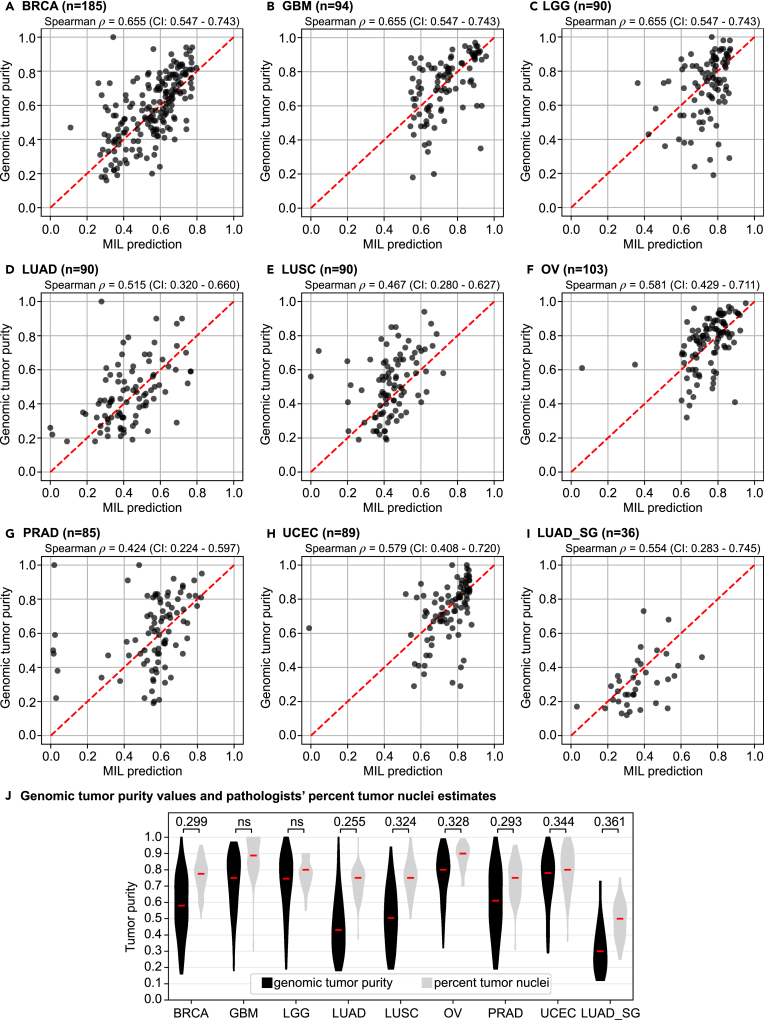
Figure 2.
The MIL model's tumor purity predictions correlate significantly with genomic tumor purity values
(A–I) A scatterplot of genomic tumor purity versus the MIL model's prediction is given for only tumor samples in the test set of each cohort: (A) BRCA, (B) GBM, (C) LGG, (D) LUAD, (E) LUSC, (F) OV, (G) PRAD, (H) UCEC, and (I) LUAD_SG. Correlation coefficients with 95% CIs are given at the top of each plot. Note that the red dotted line in each plot shows the diagonal (i.e., y = x line). All data points would align on the diagonal line in case of zero prediction error.
(J) Violin plots summarize genomic tumor purity values and pathologists' percentage tumor nuclei estimates in the test set of each cohort. Correlation coefficients are given at the top. Red lines show median values. ns, not significant; n, the number of tumor samples.
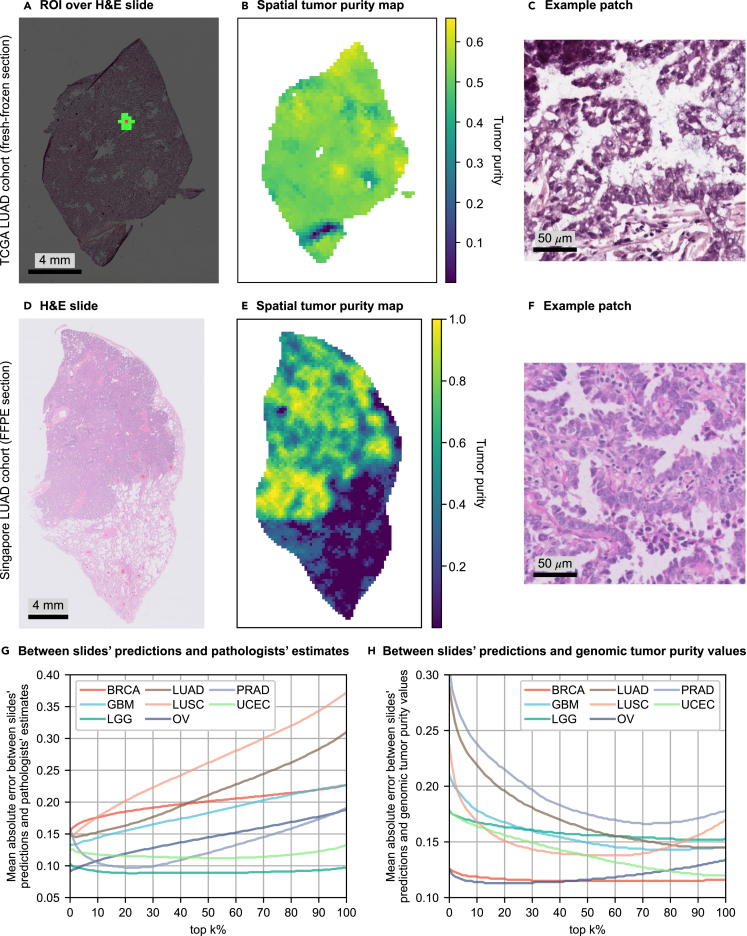
Figure 4.
Incorrect size and selection of ROI might cause overestimation in percentage tumor nuclei estimates
(A–H) For a slide of a fresh-frozen section in the TCGA LUAD cohort, (A) shows the ROI centered on a patch and consisting of 16 closest patches to that particular patch (≈1 mm2 at the specimen level). Tumor purity corresponding to the patch is predicted over the ROI. (B) The tumor purity map for all patches within the slide. Similarly, (D) shows a slide of a FFPE section in the LUAD_SG cohort, and (E) shows its corresponding tumor purity map. (C) and (F) show example patches cropped from cancerous regions in the slides shown in (A) and (D), respectively. (G and H) To investigate the effect of the size and selection of ROI on pathologists' percentage tumor nuclei estimates, we conducted error analyses over the slides' tumor purity values by gradually extending the ROI. We calculated the slide's tumor purity as the average of top-k% of the patches with the highest scores (k = 0, ···, 100) in the tumor purity map (k = 0: the patch with the highest tumor purity). In different cohorts, we plotted mean absolute error versus top-k% of the patches for error analyses between slides' predictions and pathologists' percentage tumor nuclei estimates in (G) and slides' predictions and genomic tumor purity values in (H). See also Figures S4 and S6–S10.
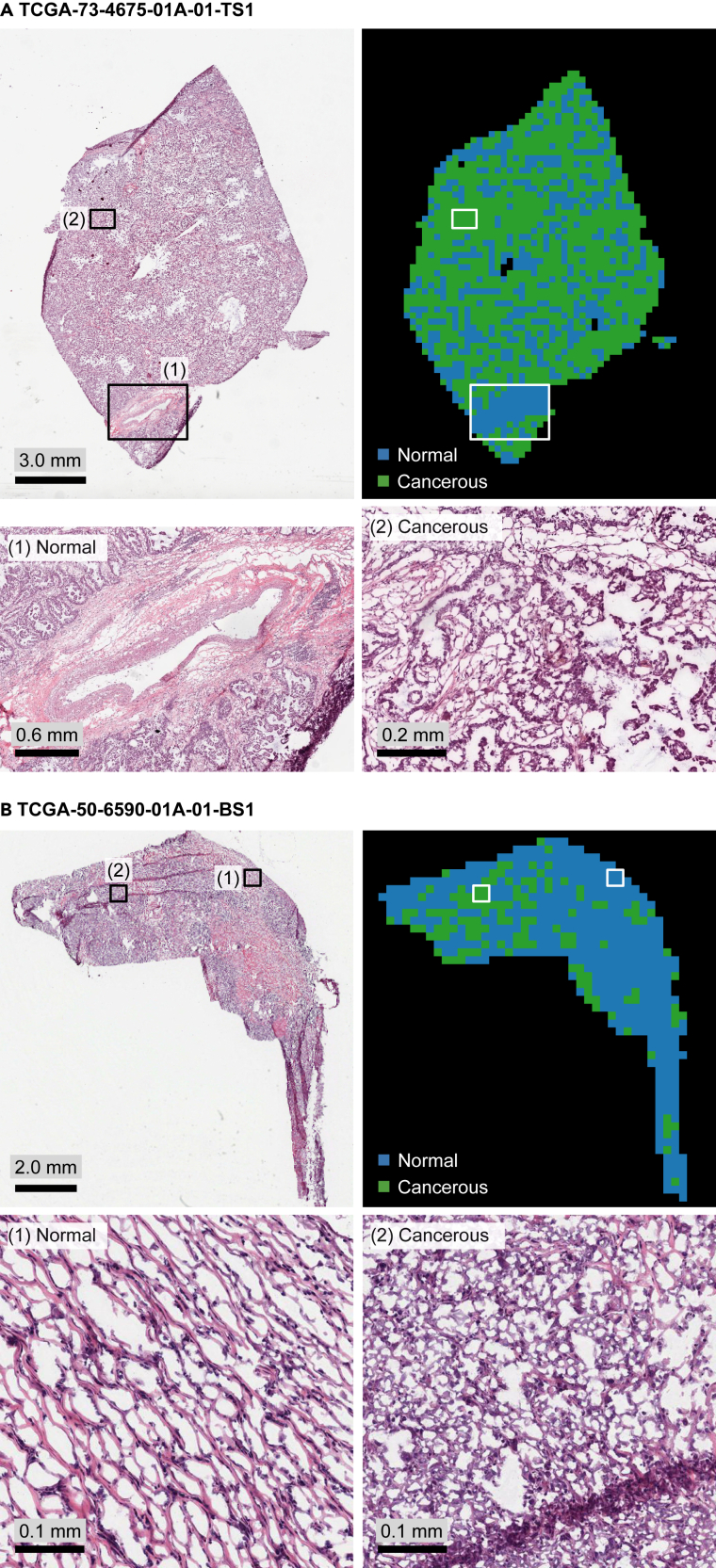
Figure 5.
Cancerous versus normal segmentation maps obtained by performing a clustering over the features extracted by the trained MIL model's feature extractor module are consistent with LUAD histopathology
(A and B) We show H&E-stained slides, color-coded segmentation maps, and example zoom-in areas for two slides in the test set of LUAD cohort: (A) TCGA-73-4675-01A-01-TS1, and (B) TCGA-50-6590-01A-01-BS1. See also supplemental experimental procedures for details.
Figure 3.
Performance analysis of MIL models
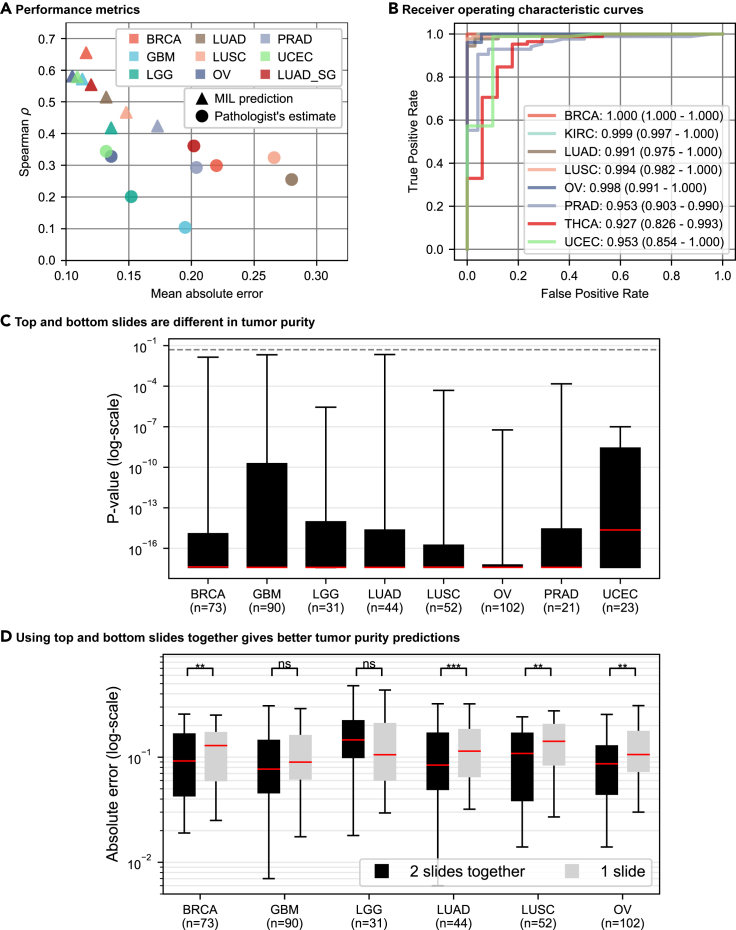
(A and B) MIL models perform better than percentage tumor nuclei estimates and successfully classify samples into tumor versus normal. (A) Spearman's correlation coefficient versus mean-absolute-error plot is given for MIL models' tumor purity predictions (represented by triangles) and pathologists' percentage tumor nuclei estimates (represented by circles) in the test sets of different cohorts (showed in different colors). MIL models' predictions achieve lower mean absolute error and higher Spearman's correlation coefficient than percentage tumor nuclei estimates. See also Tables S3 and S5. (B) ROC curve analysis over MIL models' predictions for tumor versus normal sample classification. The area under curve values with 95% CIs are given in the legend. MIL models successfully classified samples into tumor versus normal in all cohorts.
(C and D) The top and bottom slides of a tumor sample are different in tumor purity. In the test set of each cohort, for a tumor sample having top and bottom slides, we conducted two experiments. (C) The trained MIL model's predictions from the top and bottom slides of a sample are statistically compared using Wilcoxon signed-rank test.53 Each box plot summarizes the p values obtained in a cohort. For at least 95% of the samples in each cohort, the top and bottom slides are significantly different (p < 0.05) in tumor purity. The dashed line shows p = 0.05. See also Table S6. (D) For each sample, the absolute error between genomic tumor purity value and the MIL model's prediction using both slides and the expected value of absolute errors between genomic tumor purity value and the MIL model's predictions over individual slides are calculated. Box plots summarize the absolute errors in two approaches. They are statistically compared using Wilcoxon signed-rank test,53 and the results are presented on top of the plots such that p > 0.05 (ns, not significant), ∗p ≤ 0.05, ∗∗p ≤ 0.01, and ∗∗∗p ≤ 0.001. See also Table S7. Whiskers show 5th and 95th percentiles, and red lines show median values. n, number of tumor samples with two slides.
Results
In this study, there were 10 different TCGA cohorts and a local Singapore cohort. Each TCGA cohort had more than 400 patients, and the Singapore cohort had 179 lung adenocarcinoma patients, such that each patient had both histopathology slides and corresponding genomic sequencing data (Table 1, see also Tables S1 and S2). The histopathology slides in each cohort were randomly segregated at the patient level into training, validation, and test sets (Figures S1 and S2). We trained our MIL model on the training set and chose the best set of model weights based on validation set performance. Finally, we evaluated the performance of our trained MIL model on the data of completely unseen patients in the hold-out test set. Each patient in the test set was like a new patient walking into the clinic.54
Table 1.
The TCGA and Singapore cohorts
| Tumor samples |
Normal samples |
|||||
|---|---|---|---|---|---|---|
| Cohorts | Train | Validation | Test | Train | Validation | Test |
| BRCA | 559 | 185 | 185 | 76 | 27 | 30 |
| GBM | 285 | 95 | 94 | 0 | 0 | 0 |
| KIRC | 261 | 85 | 89 | 220 | 71 | 73 |
| LGG | 273 | 91 | 90 | 0 | 0 | 0 |
| LUAD | 266 | 90 | 90 | 101 | 37 | 33 |
| LUSC | 273 | 90 | 90 | 132 | 41 | 47 |
| OV | 310 | 103 | 103 | 53 | 13 | 18 |
| PRAD | 258 | 85 | 85 | 72 | 15 | 24 |
| THCA | 258 | 85 | 85 | 48 | 18 | 17 |
| UCEC | 270 | 90 | 89 | 18 | 4 | 10 |
| LUAD_SG | 107 | 36 | 36 | 0 | 0 | 0 |
In each cohort, a patient has only one tumor sample and one matching normal sample, if available. The numbers of tumor and matching normal samples in training, validation, and test sets are presented for each cohort. The data are segregated at the patient level. See also Tables S1 and S2, Figures S1–S3, and Note S1.
MIL models' tumor purity predictions correlate significantly with genomic tumor purity values
Our models' performance in 10 different TCGA cohorts was evaluated by correlation analyses between genomic tumor purity values obtained from ABSOLUTE19 and our MIL models' predictions. The performance metric was Spearman's rank correlation coefficient.
We obtained significant correlations (p < 0.05) in eight cohorts, namely breast invasive carcinoma (BRCA), glioblastoma multiforme (GBM), brain lower grade glioma (LGG), lung adenocarcinoma (LUAD), lung squamous cell carcinoma (LUSC), ovarian serous cystadenocarcinoma (OV), prostate adenocarcinoma (PRAD), and uterine corpus endometrial carcinoma (UCEC) (Figures 2A–2H and Table S3). While the minimum Spearman's = 0.418 (p = 4.1 × 10−5; 95% confidence interval [CI], 0.226–0.574) was in the LGG cohort, the maximum Spearman's = 0.655 (p = 4.6 × 10−24; 95% CI, 0.547–0.743) was in the BRCA cohort. We compared our MIL models'’ predictions with tumor purity values obtained from ESTIMATE32 as well and observed similar performance (Table S4).
We repeated the correlation analyses between genomic tumor purity values and pathologists' percentage tumor nuclei estimates (Figure 2J and Table S3). While the minimum Spearman's = 0.240 (p = 2.7 × 10−2; 95% CI, 0.009–0.446) was in the thyroid carcinoma (THCA) cohort, the maximum Spearman's = 0.344 (p = 9.8 × 10−4; 95% CI, 0.139–0.531) was in the UCEC cohort. There was no significant correlation in the GBM and LGG cohorts. Hence, the minimum correlation with MIL predictions ( = 0.418 in the LGG cohort) was higher than the maximum correlation with pathologists' percentage tumor nuclei estimates ( = 0.344 in the UCEC cohort). This implies that MIL predictions are more consistent with genomic tumor purity values than the pathologists' percentage tumor nuclei estimates.
Moreover, we conducted statistical tests on correlation coefficients to compare our MIL models' predictions and pathologists' percentage tumor nuclei estimates. We used the Fisher's z transformation-based method of Meng et al.55 Two methods were compared only when there was a significant correlation for both methods in a cohort (Table S3). MIL predictions were significantly better than pathologists' estimates in all cohorts except LUSC and PRAD. For these cohorts, two methods performed on par (pcomp = 1.7 × 10−1 > 0.05 for the LUSC and pcomp = 2.0 × 10−1 > 0.05 for the PRAD) in the test sets.
MIL models' predictions have lower mean absolute error than percentage tumor nuclei estimates
Apart from Spearman's correlation coefficients, we also checked the mean absolute errors between genomic tumor purity values and MIL models' predictions, and between genomic tumor purity values and pathologists' percentage tumor nuclei estimates (Table S5).
In the analyses of MIL predictions, the minimum and maximum mean-absolute-error values of = 0.105 (standard deviation = 0.091) and = 0.173 ( = 0.154) were obtained in the OV cohort and the PRAD cohort, respectively. On the other hand, in the analyses of pathologists' percentage tumor nuclei estimates, the minimum and maximum mean-absolute-error values of = 0.132 ( = 0.124) and = 0.280 ( = 0.151) were obtained in the UCEC cohort and the LUAD cohort, respectively. In all cohorts, pathologists' estimates were generally higher than genomic tumor purity values (Figure 2J).
Similar to our comparison in correlation analyses, we compared two methods based on absolute errors in the test sets of different cohorts. We used the Wilcoxon signed-rank test53 on absolute error values for tumor samples in the test sets (Table S5). Absolute error values in MIL predictions were significantly lower than those in pathologists' percentage tumor nuclei estimates in all cohorts except the LGG cohort. Two methods performed similarly (pcomp = 5.4 × 10−2 > 0.05) in the test set of the LGG cohort.
Figure 3A summarizes correlation and absolute error analyses. We observed that MIL predictions had lower mean absolute error and higher Spearman's correlation coefficient than pathologists' percentage tumor nuclei estimates.
MIL model predicts tumor purity from H&E-stained slides of FFPE sections in the Singapore cohort
Our MIL models successfully predicted tumor purity from H&E-stained digital histopathology slides of fresh-frozen sections in different TCGA cohorts. Besides, we evaluated their performance on slides of formalin-fixed paraffin-embedded (FFPE) sections in a local Singapore cohort consisting of 179 lung adenocarcinoma patients (see Note S1 for details). Similar to TCGA cohorts, we segregated data at the patient level (Table 1 and Figure S3).
We used transfer learning and initialized the model with the weights of the MIL model trained on the TCGA LUAD cohort. Then, we froze the weights of all layers in the network except the first convolutional layer in the feature extractor module (Figure 1A). This helped the network adapt the first layer weights to learn the tissue morphology in FFPE sections, which were different from fresh-frozen sections (Figure S4). Note that, while the FFPE method preserves morphology better and is the routine in histopathology, the fresh-frozen method preserves nucleic acids better and is preferred for molecular analysis.56
Similar to the performance in the TCGA LUAD cohort, we obtained a Spearman's = 0.554 (p = 4.6 × 10−4; 95% CI, 0.283–0.745) and the mean absolute error of = 0.120 ( = 0.091) in the test set of the Singapore LUAD (LUAD_SG) cohort (Figures 2I and 3A). There were substantial differences between the TCGA and LUAD_SG cohorts, such as tissue preservation method (fresh-frozen versus FFPE) and ancestry of patients (European versus East Asian). However, our MIL model successfully predicted tumor purity from slides of FFPE sections using transfer learning with minimal training only in the first convolutional layer of the feature extractor module. The results suggested that our MIL models learned robust features for tumor purity prediction tasks at the higher levels of the network. We also checked the performance of the TCGA LUAD model directly on the LUAD_SG cohort used as an external validation set (Figure S5). Nevertheless, we did not get a significant correlation ( = 0.141; p = 0.06 > 0.05), which highlighted the necessity of adapting the weights of the first layer in feature extractor to FFPE slides using transfer learning.
For pathologists' estimates, we obtained a Spearman's = 0.361 (p = 3.0 × 10−2; 95% CI, 0.029–0.644) and the mean absolute error of = 0.202 ( = 0.105) in the test set of the LUAD_SG cohort (Figure 2J). We statistically compared the MIL model's predictions and percentage tumor nuclei estimates. While the difference was not significant (pcomp,ρ = 2.3 × 10−1 > 0.05) in terms of correlation coefficient, it was significant (pcomp,abs = 7.3 × 10−4 < 0.05) in terms of absolute error.
Tumor purity varies spatially within a sample: Top and bottom slides of a sample are different in tumor purity
Intra-tumor heterogeneity is a well-known phenomenon in solid cancers.57, 58, 59, 60, 61 It results in therapeutic failure and drug resistance.62 We checked whether it is observable from tumor purity predictions of the trained MIL model on the top and bottom slides of a sample. For each slide of a tumor sample with both top and bottom slides in a cohort, 100 bags are created from the slide's patches and predictions are obtained from the trained MIL model. Then, the predictions of two slides are statistically compared using the Wilcoxon signed-rank test.53
Figure 3C shows the box plot of p values obtained from the statistical tests in each cohort's test set. There is a significant difference between the MIL predictions on the top and bottom slides of the same tumor sample. In all cohorts, at least 75% of samples have p value p < 1.0 × 10−8 and at least 95% of samples have p value p < 0.05. Hence, we conclude that there is a variation in tumor purity between the top and bottom sections of a tumor sample; i.e., tumor purity varies spatially within the sample.
The degree of spatial variation in tumor purity is different for different cancer types (Table S6). The UCEC, LGG, and GBM cohorts had the lowest mean absolute differences (μdabs) between top and bottom slides' predictions (μdabs ≤ 0.090); i.e., they were the most spatially homogeneous cancers among all cohorts. On the other hand, the PRAD cohort had the highest mean absolute difference (μdabs = 0.144); i.e., it was the most spatially heterogeneous cancer in tumor purity.
Predicting a sample's tumor purity using both top and bottom slides is better than using only one slide
We checked if there is a significant difference between predicting a sample's tumor purity by using both slides (top and bottom) and using only one slide. For a tumor sample with two slides in a cohort, let psmpl be genomic tumor purity value of the sample; be tumor purity prediction obtained from trained MIL model by using both of the slides together; and be tumor purity predictions obtained from trained MIL model for individual slides. We compared the absolute error of sample-level prediction esmpl = | - psmpl| and the expected value of absolute errors of slide-level predictions esld = 0.5 ∗ (| - psmpl| + | - psmpl|). We used the Wilcoxon signed-rank test53 on the difference of esmpl − esld (Table S7). Note that the PRAD (n = 21) and UCEC (n = 23) cohorts were excluded from this study due to few samples with two slides.
In the test sets of BRCA, LUAD, LUSC, and OV cohorts, using both slides for tumor purity prediction gave better results in terms of absolute error (Figure 3D). However, in the test sets of GBM and LGG cohorts, there was no significant difference using both slides or one slide alone. Indeed, this is not surprising since they had the lowest mean absolute differences between the slides' predictions (Table S6); i.e., the most spatially homogeneous tumors. In fact, when both slides are the same, sample-level prediction and slide predictions would be the same.
We conclude that predicting a sample's tumor purity using both the top and bottom slides together is better than using only one of them whenever possible.
Spatial tumor purity map analysis reveals the probable cause of pathologists' high percentage tumor nuclei estimates
Pathologists' percentage tumor nuclei estimates were generally higher than genomic tumor purity values for all TCGA cohorts in our analysis (Figure 2J and see also Figures S1 and S2). Previous studies also stated that,5,13 but the reasons remain unclear. We hypothesized that incorrect size and selection of ROI might be the cause. We obtained tumor purity maps by our trained MIL models in different TCGA cohorts and conducted error analysis over them to test our hypothesis.
We followed the same procedure as in Smits et al.6 to simulate pathologists' percentage tumor nuclei estimation. Tumor purity is predicted over an ROI of 1 mm × 1 mm around each patch in a slide, which corresponds to 16 patches at 20× zoom level (each patch is around 256 μm × 256 μm at the specimen level) (Figure 4A). Then, the predicted value is assigned to the patch in the tumor purity map (Figure 4B). We also obtained tumor purity maps for slides in the Singapore cohort (Figures 4D, 4E, and S6–S10).
We observed that a tumor purity map shows variation within the slide, which implies that ROI selection is crucial in pathologists' percentage tumor nuclei estimation. Since tumor purity was higher in pathologists' percentage tumor nuclei estimates, we investigated whether pathologists might have selected high tumor content regions over the slides for percentage tumor nuclei estimation. The highest prediction in a slide's tumor purity map was used as the slide's tumor purity value. Then, error analyses were conducted over the slides' tumor purity values compared with pathologists' percentage tumor nuclei estimates and genomic tumor purity values. The error analyses were repeated by gradually extending the ROI such that a slide's tumor purity was calculated as the average of top-k% of the patches with the highest scores (k = 0, ···, 100) in the slide's tumor purity map.
We discovered that the mean absolute error between the slides' predictions and pathologists' percentage tumor nuclei estimates increases as we extend the ROI to cover the lower tumor purity regions (Figure 4G). These observations suggested that pathologists may tend to select high-tumor-content regions to estimate percentage tumor nuclei. The LGG and UCEC cohorts may look exceptional with almost constant mean-absolute-error plots. However, this is expected since these two cohorts' samples have high genomic tumor purity values (Figure S1), so the variation within the slides is very low. The PRAD cohort's plot also has a different pattern than the others. It has an initial decrease and an increase in the later stages, emphasizing the importance of the ROI size. The pathologists may need to analyze a bigger ROI depending on the morphology of the tissue origin to reach a certain nuclei count while estimating percentage tumor nuclei. The PRAD may be one of them due to the glandular structure of the prostate.
Furthermore, as the ROI grows, the mean absolute error between the slides' predictions and genomic tumor purity values decreases (Figure 4H). Indeed, this is expected since our MIL models converge to their original performance of prediction over the whole slide (Figure 2). It is even more evident in the LUSC and OV cohorts. The error decreases initially but increases later since our MIL models underestimated the tumor purity compared with genomic tumor purity values in these cohorts.
MIL model learns discriminant features for cancerous versus normal tissue histology
We explored the capability of our MIL model's feature extractor on learning discriminant features for cancerous versus normal tissue histology while being trained on sample-level genomic tumor purity labels. For each patient having both tumor and matching normal samples, features of patches cropped over the slides of the tumor and normal samples were extracted using the trained feature extractor module of the MIL model. Then, slide-level cancerous versus normal segmentation maps were obtained by performing a hierarchical clustering over the extracted feature vectors (Figure 1C and see supplemental experimental procedures for details). The resolution of segmentation was at the patch level, and each patch was around 256 μm × 256 μm at the specimen level.
In the test set of the LUAD cohort, there were 33 patients both with tumor and matching normal samples. We constructed slide-level segmentation maps for these patients (Figure 5). We observed that segmentation maps were consistent with the LUAD histopathology during the qualitative assessment of the segmentation maps. While healthy tissue components, like blood vessels, stroma regions, and normal tissue structures, were labeled normal, regions invaded by neoplastic cells were labeled cancerous. Hence, we qualitatively validated that our MIL model learned discriminant features for cancerous versus normal tissue histology in LUAD from sample-level genomic tumor purity labels without requiring pixel-level annotations from pathologists.
MIL model successfully classifies samples into tumor versus normal
A good tumor purity predictor should be able to discriminate between tumor and normal. We checked our MIL model's performance in the tumor versus normal sample classification task. Tumor purity predictions for all samples in the test set of each cohort were obtained and a receiver operating characteristic (ROC) curve analysis was conducted. Then, the area under the ROC curve (AUC) was calculated and a 95% CI was constructed using the percentile bootstrap method.63 Note that GBM and LGG cohorts were excluded from analysis since there were no normal slides in these cohorts.
Our MIL models successfully discriminated tumor samples from normal samples in all cohorts with AUC values greater than or equal to 0.927 (Figure 3B). We got the minimum and maximum AUC values of 0.927 (95% CI, 0.826–0.993) and 1.000 (95% CI, 1.000–1.000) on the test sets of THCA and BRCA cohorts, respectively. Note that, although we did not get a strong correlation between genomic tumor purity values and MIL predictions in the test sets of kidney renal clear cell carcinoma (KIRC) and THCA cohorts, our models successfully classified samples into tumor versus normal in these cohorts.
Furthermore, we obtained an AUC value of 0.991 (95% CI, 0.975–1.000) on the test set of the LUAD cohort. Our model outperformed the classical image processing and machine-learning-based method of Yu et al.64 (AUC, 0.85) and the DNA plasma-based method of Sozzi et al.65 (AUC, 0.94). Besides, our model performed on par with the deep learning model of Coudray et al.66 (AUC, 0.993), which was trained on tumor versus normal classification, and the deep learning model of Fu et al.67 (AUC, 0.977 with 95% CI, 0.976–0.978), which was fine-tuned on pathologists' percentage tumor nuclei estimates in a transfer learning setup. However, there is one concern about the dataset preparation methods of Coudray et al.66 and Fu et al.67 They obtained the datasets by segregating data either at slide level66 or at patch level.67 These data segregation methods might lead to a severe data leakage problem, and the models' performance might be illusory.
Discussion
Accurate tumor purity estimation is crucial for high-throughput genomic analysis. It is routinely estimated by pathologists; however, pathologists' estimates suffer from inter-observer variability and do not correlate well with genomic tumor purity values. Besides, percentage tumor nuclei estimation by pathologists is tedious and time consuming. To overcome these challenges, we developed a novel MIL model with a distribution pooling filter. It predicted tumor purity from H&E-stained histopathology slides of fresh-frozen and FFPE sections in different TCGA cohorts and a Singapore cohort, respectively. The predictions were consistent with genomic tumor purity values, and they outperformed pathologists' percentage tumor nuclei estimates in the TCGA cohorts.
Hence, our MIL models can be utilized for sample selection for high-throughput genomic analysis, which will help reduce pathologists' workload and decrease inter-observer variability. Moreover, spatially resolved tumor purity maps obtained using our MIL models can substantially contribute to a better understanding of the tumor microenvironment. Lastly, our models' predictions can be used as prognostic biomarkers to stratify patients.
MIL model can pre-screen slides for genomic analysis
The current workflow for sample selection for genomic analysis includes screening slides of 8–12 sections, choosing the most appropriate slide, and, possibly, marking out a high tumor content region on the slide for macrodissection before extraction. This adds a heavy burden to pathologists' workload. To help pathologists, our MIL model can pre-screen the slides and suggest the best slide (with the highest predicted tumor purity) for high-throughput sequencing. Moreover, it can propose high-tumor-content regions over the slide for macrodissection via spatial tumor purity map, which is remarkably important for low-purity samples (for example, in lung cancers).
Furthermore, our MIL model's predictions can be used as a quality control metric to decide if a section has enough tumor content for sequencing or if a section requires deeper sequencing. This can avoid wasting the limited amount of tissue (especially in biopsy samples) in failed sequencing attempts.
Genomic tumor purity values and pathologists' percentage tumor nuclei estimates are complementary
While genomic tumor purity values have recently been recognized as accurate for downstream genomic analysis after sequencing,35 sequencing is still subject to sample selection based on pathologists' percentage tumor nuclei estimates. On the other hand, pathologists' slide reading method is inherently limited since it requires cellular-level analysis. It can give reliable results over a selected ROI, but it may not be applicable for sample-level tumor purity prediction, ideally requiring the analysis of gigapixel digital histopathology slides. Therefore, we used sample-level genomic tumor purity values as labels for training our MIL models. Now, we can use our MIL models to support pathologists for sample selection for molecular analysis by pre-screening slides and proposing ROIs for further assessment.
Spatially resolved tumor purity maps can enhance spatial omics
We obtained tumor purity maps showing the variation of tumor purity in slides using our trained MIL models (Figures 4B and 4E). They can potentially help understand the interaction of cancer cells with other tissue components (like normal epithelial, stromal, and immune cells) in the tumor microenvironment, which is a key player in tumor formation and primary determinant of therapeutic response.68,69 Furthermore, they can enhance spatial-omics technologies.70, 71, 72, 73
Weak tumor purity labels innately necessitated an MIL approach
Previous studies based on patch-based models worked on few cancer types with relatively few patients (like 10 patients46 or 64 patients47,48) since they required pixel-level annotations (rarely available). However, using genomic tumor purity values as sample-level weak labels enabled us to conduct a pan-cancer study on 10 different TCGA cohorts, where each cohort had more than 400 patients. On the other hand, unlike pixel-level annotations providing whether each cell is cancerous or normal, the genomic tumor purity of a sample tells us only the proportion of cancer cells within the sample. Therefore, training a machine learning model using weak tumor purity labels innately necessitated an MIL approach, where a sample was represented as a bag of patches from the sample's slides, and the sample's genomic tumor purity value was used as the bag's label.
The sources of error in MIL predictions
Our MIL models successfully predicted tumor purity (Figure 2). However, they slightly deviated from the genomic tumor purity values. There may be different sources of prediction errors. While some of them can be eliminated, some are inevitable.
First, we have fewer patients in our datasets (300 patients per training set) than traditional deep learning datasets containing millions of independent samples.74 Considering the complexity of cancer, our MIL models effectively captured features that distinguish cancerous versus normal. We also expect that the performance will improve with the increasing number of patients. Indeed, we obtained the best performance in our largest cohort of BRCA (559 patients in the training set).
Second, our MIL model uses histopathology slides from the top and bottom sections of the tumor portion. We have already shown the variation in tumor purity between the top and bottom sections of the tumor samples. Thus, for samples with only one slide, the prediction error is expected to be higher.
Third, we checked if necrosis regions inside the slides affect our MIL models' performances. For each cohort (except the LGG cohort, in which all samples have percentage necrosis of 0), Spearman's correlation coefficients between absolute errors in MIL predictions and percentage necrosis values are calculated in the test set (Table S8). There is no significant correlation (p > 0.05) in any cohorts except the LUSC cohort, in which we observe a low correlation of 0.253 (p = 1.6 × 10−2; 95% CI, 0.062, 0.432). Overall, it seems that our models can handle necrosis regions well.
Last, our model's predictions are based on morphology in H&E-stained histopathology slides. However, genomic tumor purity values were based on DNA data, and all the effects of genetic changes (so the genomic tumor purity changes) may not be observable from the slides due to the selective dyeing characteristics of H&E staining. Even some genetic changes may not manifest in morphology.75
Why do MIL predictions perform better than percentage tumor nuclei estimates?
Compared with the percentage tumor nuclei estimates by pathologists, our MIL models' predictions gave a higher correlation and lower mean absolute error with genomic tumor purity values (Figure 3A). One of the primary reasons for this superiority is that the MIL models were trained directly on genomic tumor purity values, which enabled the MIL models to learn associated features.
Another reason might be that pathologists concentrate more on tumor cells than infiltrating normal cells within the tumor, which may result in missed normal tissue components. Moreover, cancer cells are usually enlarged. They occupy more space than normal tissue components, stromal cells, and infiltrating lymphocytes, which may create an implication of high tumor content.18 Pathologists may fail to incorporate this effect in their estimates correctly and may overestimate percentage tumor nuclei. Indeed, this was the case in the cohorts we analyzed (Figures 2J, S1, and S2).
Finally, while our MIL models predict tumor purity over the whole slide, pathologists estimate the percentage tumor nuclei by analyzing some selected ROI over the slide. Therefore, the size and selection of the ROI might cause the overestimation in pathologists' percentage tumor nuclei estimates (Figure 4G).
Limitations and future work
Our MIL models, by design, apply to any tumor sample with H&E-stained histopathology slides. We tested them on tumor samples with a broad range of tumor purity values. However, testing them on samples with percentage tumor nuclei lower than TCGA threshold would strengthen the applicability of our MIL models. It is reserved for future work.
We evaluated our MIL models on hold-out test sets to simulate real-world clinical workflow and obtained successful results. Besides, our analysis on the LUAD_SG cohort using transfer learning with minimal training for domain adaptation showed that our MIL models learned robust features for tumor purity prediction tasks. However, we could not validate our models on external cohorts due to differences between fresh-frozen and FFPE tissue preservation methods, which might further consolidate their robustness.
We qualitatively validated our spatially resolved tumor purity maps. Quantitative validation of them using spatial-omics technologies is reserved for future work, which requires recruitment of a prospective cohort, conducting spatial-omics and image analysis, and evaluating purity maps obtained from the MIL model against spatial-omics.
Lastly, our MIL models are deep learning based, and deep learning algorithms perform better with more data. Training of the models with larger cohorts would help to improve the model performance by better capturing patient-to-patient variations.
Experimental procedures
Resource availability
Lead contact
Further information and requests for resources should be directed to and will be fulfilled by the lead contact, Wing-Kin Sung (ksung@comp.nus.edu.sg).
Materials availability
This study did not generate new unique reagents.
Datasets
We downloaded H&E-stained histopathology slides of fresh-frozen sections in 10 different cohorts in TCGA (Table 1). We selected these cohorts since they have more than 400 patients with both histopathology slides and corresponding genomic sequencing data in TCGA. Each patient had a tumor sample, and some patients also had matching normal samples.
In TCGA, primary tumor samples and matching normal samples (adjacent non-neoplastic solid tissue or blood) were collected at the Tissue Source Sites (TSSs) from patients who had received no prior treatment (chemotherapy or radiotherapy) for their disease. Collected samples were frozen and shipped overnight to the Biospecimen Core Resource (BCR) for TCGA while maintaining a temperature less than −180°C.76
At the BCR, each frozen sample was cut into portions.77 Then, two glass slides (sometimes only one) were prepared by cutting sections 4–6 μm thick from the top and bottom of a portion78 and staining with H&E.79 Based on the information from the BCR (via personal communication), these slides were scanned at 40× magnification using an Aperio XT slide scanner. A board-certified pathologist reviewed the slides. Upon passing pathology review, the remaining portion without any tumor enrichment was sent for genomic analysis.80 In other words, the (top and bottom) slides and the portion sent for genomic analysis were immediate neighbors.
During review (personal communication), a pathologist estimated (1) percentage tumor nuclei and percentage of all other nuclei, which add up to 100%; and (2) percentage cellular tumor, percentage normal, percentage stroma, and percentage necrosis, which add up to 100%. Percentage tumor nuclei in each slide of a frozen tumor section was estimated by evaluating at least 10 specimen fields (excluding necrosis regions) via the digital slide viewer. Tumor portions with percentage tumor nuclei of ≥60% and percentage necrosis of ≤20% were accepted to the study and sent for genomic analysis.76 Besides, pathologists confirmed from slides of frozen normal sections that the adjacent normal tissues (if available) were free of tumor cells.
We also collected H&E-stained histopathology slides of FFPE sections in an East Asian cohort consisting of 179 lung adenocarcinoma patients in Singapore. In the Singapore cohort, only one slide was prepared for each tumor sample from the top section of the tissue used for sequencing, and there were no normal samples. All the slides of FFPE sections were prepared, stained, and scanned at 40× magnification using The Philips IntelliSite Pathology Solution (Koninklijke Philips, The Netherlands) in the same laboratory in Singapore.
In each cohort, we randomly segregated the data at the patient level (i.e., slides from the same patient should be in the same set) into training (60%), validation (20%), and test (20%) sets (Table 1), which had similar tumor purity distributions (Figures S1 and S2). Note that segregating data at the patient level is crucial to prevent data leakage while training machine learning models.54 The training set was used to train the machine learning model, the validation set was used to choose the best model, and the test set was held out as unseen data for evaluation of the best model. The list of patients and slides in each set are given in Document S2.
MIL model
Our novel MIL model consists of three modules: feature extractor module, MIL pooling filter, and bag-level representation transformation module (Figure 1A). We use neural networks to implement the feature extractor module and the bag-level representation transformation module to parameterize the learning process fully (see supplemental experimental procedures for details). We use our novel distribution pooling filter as the MIL pooling filter. It is more expressive than standard pooling filters (like mean and maximum pooling) regarding the amount of information captured while obtaining bag-level representations.81 Given a bag of patches, the feature extractor module extracts a feature vector for each patch inside the bag. Then, thanks to its superiority, the distribution pooling filter obtains a strong bag-level representation by estimating the marginal distributions of the extracted features. Finally, the bag-level representation transformation module predicts tumor purity. This system of neural network modules is trained end-to-end using samples' genomic tumor purity values as labels.
Training of MIL models
To prepare machine learning datasets, tissue regions inside histopathology slides were detected by applying OTSU thresholding. Over the tissue regions, non-overlapping 512 × 512 patches at 20× zoom level (specimen-level pixel size, 0.5 μm × 0.5 μm) were cropped.
During training, we used a bag of patches cropped from a sample's top and bottom slides as the input and the sample's tumor purity value obtained from genomic sequencing data by ABSOLUTE19 as the ground-truth label (Figure S1). At each epoch, one bag per sample is created on the fly by randomly selecting 200 patches from the sample's patches. We also used the matching normal samples whenever available to enable our model to capture the information related to normal tissue histology. We assigned a tumor purity value of 0.0 to a matching normal sample as the ground-truth label. Note that there were no normal samples in the Singapore cohort.
We initialized the models' weights randomly and trained them end-to-end using the ADAM optimizer with a learning rate of 0.0001 and L2 regularization on the weights with a weight decay of 0.0005. The batch size was 1. We used absolute error as the loss function and employed early stopping based on loss in the validation set to avoid overfitting.
Predicting tumor purity of a sample
We created 100 bags for each sample in the test set and obtained tumor purity predictions from the trained model. Each bag was created by randomly selecting 200 patches from the available patches cropped from the sample's (top and bottom) slides. We used the average of 100 predictions as the sample's tumor purity prediction during performance evaluation.
Statistical analysis
We obtained 95% CIs for Spearman's rank correlation coefficients and area under the ROC curves using the percentile bootstrap method.63 To compare the performance of two methods (our MIL models' predictions and pathologists' percentage tumor nuclei estimates), we used the Fisher's z transformation-based method of Meng et al.55 on Spearman's rank correlation coefficients and Wilcoxon signed-rank test53 on absolute error values.
All statistical tests were two sided and statistical significance was considered when p < 0.05. We used scipy.stats (v1.4.1) python library for statistical tests.
Acknowledgments
This work is partly supported by the Biomedical Research Council of the Agency for Science, Technology and Research, Singapore, and the National University of Singapore, Singapore. We thank Jay Bowen (Principal Investigator at the BCR) for his detailed explanations about how the slides are prepared, stained, and scanned at the BCR for TCGA and providing detailed information about the pathology review process of the slides at the BCR for sample selection for downstream analyses. We thank Valerie Yang for constructive comments on the manuscript.
Author contributions
M.U.O., W.K.S., and H.K.L. conducted the machine learning study. J.C., E.R., A.N.K., J.J.S.A., W.Z., and A.J.S. performed genomic analysis. D.S.W.T. designed the clinical study of the Singapore cohort. A.T. and X.M.C. selected the cases in the Singapore cohort and conducted the histopathology review. A.J. retrieved the slides and estimated percentage tumor nuclei values. S.Y.H. scanned the slides and exported digital slide images. T.K.H.L. reviewed the slides and contributed to the methods discussion and manuscript review.
Declaration of interests
The authors declare no competing interests.
Published: February: 11, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.patter.2021.100399.
Supplemental information
Data and code availability
-
•
All TCGA datasets are publicly available. Manifest files are provided with original code to download H&E-stained digital histopathology slides using GDC Data Transfer Tool. Genomic tumor purity values were downloaded from https://gdc.cancer.gov/about-data/publications/pancanatlas under filename TCGA_mastercalls.abs_ tables_JSedit.fixed.txt. They are also given in Data S1. For the Singapore cohort, genomic tumor purity values and representative histological images are publicly available from OncoSG (https://src.gisapps.org/OncoSG/) under dataset Lung Adenocarcinoma (GIS, 2019).
-
•
All original code has been deposited at Zenodo under the https://doi.org/10.5281/zenodo.5606981 and is publicly available as of the date of publication. The repository provides a detailed step-by-step explanation, from downloading H&E-stained digital histopathology slides to obtaining spatially resolved tumor purity maps (SRTPMs).
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- 1.Schuster S.C. Next-generation sequencing transforms today’s biology. Nat. Methods. 2008;5:16–18. doi: 10.1038/nmeth1156. [DOI] [PubMed] [Google Scholar]
- 2.Xuan J., Yu Y., Qing T., Guo L., Shi L. Next-generation sequencing in the clinic: promises and challenges. Cancer Lett. 2013;340:284–295. doi: 10.1016/j.canlet.2012.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jennings L.J., Arcila M.E., Corless C., Kamel-Reid S., Lubin I.M., Pfeifer J., Temple-Smolkin R.L., Voelkerding K.V., Nikiforova M.N. Guidelines for validation of next-generation sequencing–based oncology panels: a joint consensus recommendation of the Association for Molecular Pathology and College of American Pathologists. J. Mol. Diagn. 2017;19:341–365. doi: 10.1016/j.jmoldx.2017.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Whiteside T. The tumor microenvironment and its role in promoting tumor growth. Oncogene. 2008;27:5904–5912. doi: 10.1038/onc.2008.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aran D., Sirota M., Butte A.J. Systematic pan-cancer analysis of tumour purity. Nat. Commun. 2015;6:1–12. doi: 10.1038/ncomms9971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Smits A.J., Kummer J.A., De Bruin P.C., Bol M., Van Den Tweel J.G., Seldenrijk K.A., Willems S.M., Offerhaus G.J.A., De Weger, et al. The estimation of tumor cell percentage for molecular testing by pathologists is not accurate. Mod. Pathol. 2014;27:168–174. doi: 10.1038/modpathol.2013.134. [DOI] [PubMed] [Google Scholar]
- 7.Kim J., Park W.-Y., Kim N.K., Jang S.J., Chun S.-M., Sung C.-O., Choi J., Ko Y.-H., Choi Y.-L., Shim H.S., et al. Good laboratory standards for clinical next-generation sequencing cancer panel tests. J. Pathol. Transl. Med. 2017;51:191. doi: 10.4132/jptm.2017.03.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Patel N.M., Jo H., Eberhard D.A., Yin X., Hayward M.C., Stein M.K., Hayes D.N., Grilley-Olson J.E. Improved tumor purity metrics in next-generation sequencing for clinical practice: the integrated interpretation of neoplastic cellularity and sequencing results (IINCaSe) approach. Appl. Immunohistochem. Mol. Morphol. 2019;27:764–772. doi: 10.1097/PAI.0000000000000684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Elloumi F., Hu Z., Li Y., Parker J.S., Gulley M.L., Amos K.D., Troester M.A. Systematic bias in genomic classification due to contaminating non-neoplastic tissue in breast tumor samples. BMC Med. Genomics. 2011;4:1–12. doi: 10.1186/1755-8794-4-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Isella C., Terrasi A., Bellomo S.E., Petti C., Galatola G., Muratore A., Mellano A., Senetta R., Cassenti A., Sonetto C., et al. Stromal contribution to the colorectal cancer transcriptome. Nat. Genet. 2015;47:312–319. doi: 10.1038/ng.3224. [DOI] [PubMed] [Google Scholar]
- 11.Zhang W., Feng H., Wu H., Zheng X. Accounting for tumor purity improves cancer subtype classification from DNA methylation data. Bioinformatics. 2017;33:2651–2657. doi: 10.1093/bioinformatics/btx303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rhee J.-K., Jung Y.C., Kim K.R., Yoo J., Kim J., Lee Y.-J., Ko Y.H., Lee H.H., Cho B.C., Kim T.-M. Impact of tumor purity on immune gene expression and clustering analyses across multiple cancer types. Cancer Immunol. Res. 2018;6:87–97. doi: 10.1158/2326-6066.CIR-17-0201. [DOI] [PubMed] [Google Scholar]
- 13.Haider S., Tyekucheva S., Prandi D., Fox N.S., Ahn J., Xu A.W., Pantazi A., Park P.J., Laird P.W., Sander C., et al. Systematic assessment of tumor purity and its clinical implications. JCO Precis. Oncol. 2020;4:995–1005. doi: 10.1200/PO.20.00016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cheng J., He J., Wang S., Zhao Z., Yan H., Guan Q., Li J., Guo Z., Ao L. Biased influences of low tumor purity on mutation detection in cancer. Front. Mol. Biosci. 2020;7:533196. doi: 10.3389/fmolb.2020.533196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang C., Cheng W., Ren X., Wang Z., Liu X., Li G., Han S., Jiang T., Wu A. Tumor purity as an underlying key factor in glioma. Clin. Cancer Res. 2017;23:6279–6291. doi: 10.1158/1078-0432.CCR-16-2598. [DOI] [PubMed] [Google Scholar]
- 16.Mao Y., Feng Q., Zheng P., Yang L., Liu T., Xu Y., Zhu D., Chang W., Ji M., Ren L., et al. Low tumor purity is associated with poor prognosis, heavy mutation burden, and intense immune phenotype in colon cancer. Cancer Manag. Res. 2018;10:3569. doi: 10.2147/CMAR.S171855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gong Z., Zhang J., Guo W. Tumor purity as a prognosis and immunotherapy relevant feature in gastric cancer. Cancer Med. 2020;9:9052–9063. doi: 10.1002/cam4.3505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mikubo M., Seto K., Kitamura A., Nakaguro M., Hattori Y., Maeda N., Miyazaki T., Watanabe K., Murakami H., Tsukamoto T., et al. Calculating the tumor nuclei content for comprehensive cancer panel testing. J. Thorac. Oncol. 2020;15:130–137. doi: 10.1016/j.jtho.2019.09.081. [DOI] [PubMed] [Google Scholar]
- 19.Carter S.L., Cibulskis K., Helman E., McKenna A., Shen H., Zack T., Laird P.W., Onofrio R.C., Winckler W., Weir B.A., et al. Absolute quantification of somatic DNA alterations in human cancer. Nat. Biotechnol. 2012;30:413–421. doi: 10.1038/nbt.2203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Oesper L., Mahmoody A., Raphael B.J. Theta: inferring intra-tumor heterogeneity from high-throughput DNA sequencing data. Genome Biol. 2013;14:R80. doi: 10.1186/gb-2013-14-7-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen H., Bell J.M., Zavala N.A., Ji H.P., Zhang N.R. Allele- specific copy number profiling by next-generation DNA sequencing. Nucleic Acids Res. 2015;43:e23. doi: 10.1093/nar/gku1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yu G., Zhang B., Bova G.S., Xu J., Shih I.-M., Wang Y. BACOM: in silico detection of genomic deletion types and correction of normal cell contamination in copy number data. Bioinformatics. 2011;27:1473–1480. doi: 10.1093/bioinformatics/btr183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang B., Hou X., Yuan X., Shih I.-M., Zhang Z., Clarke R., Wang R.R., Fu Y., Madhavan S., Wang Y., et al. AISAIC: a software suite for accurate identification of significant aberrations in cancers. Bioinformatics. 2014;30:431–433. doi: 10.1093/bioinformatics/btt693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yuan X., Bai J., Zhang J., Yang L., Duan J., Li Y., Gao M. CONDEL: detecting copy number variation and genotyping deletion zygosity from single tumor samples using sequence data. IEEE/ACM Trans. Comput. Biol. Bioinform. 2018;17:1141–1153. doi: 10.1109/TCBB.2018.2883333. [DOI] [PubMed] [Google Scholar]
- 25.Yuan X., Yu J., Xi J., Yang L., Shang J., Li Z., Duan J. CNV_IFTV: an isolation forest and total variation-based detection of CNVs from short-read sequencing data. IEEE/ACM Trans. Comput. Biol. Bioinform. 2019;18:539–549. doi: 10.1109/TCBB.2019.2920889. [DOI] [PubMed] [Google Scholar]
- 26.Van Loo P., Nordgard S.H., Lingjærde O.C., Russnes H.G., Rye I.H., Sun W., Weigman V.J., Marynen P., Zetterberg A., Naume B., et al. Allele-specific copy number analysis of tumors. Proc. Natl. Acad. Sci. U S A. 2010;107:16910–16915. doi: 10.1073/pnas.1009843107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Andor N., Harness J.V., Mueller S., Mewes H.W., Petritsch C. EXPANDS: expanding ploidy and allele frequency on nested subpopulations. Bioinformatics. 2014;30:50–60. doi: 10.1093/bioinformatics/btt622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Favero F., Joshi T., Marquard A.M., Birkbak N.J., Krzystanek M., Li Q., Szallasi Z., Eklund A.C. Sequenza: allele-specific copy number and mutation profiles from tumor sequencing data. Ann. Oncol. 2015;26:64–70. doi: 10.1093/annonc/mdu479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Su X., Zhang L., Zhang J., Meric-Bernstam F., Weinstein J.N. PurityEst: estimating purity of human tumor samples using next-generation sequencing data. Bioinformatics. 2012;28:2265–2266. doi: 10.1093/bioinformatics/bts365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Larson N.B., Fridley B.L. PurBayes: estimating tumor cellularity and subclonality in next-generation sequencing data. Bioinformatics. 2013;29:1888–1889. doi: 10.1093/bioinformatics/btt293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yuan X., Li Z., Zhao H., Bai J., Zhang J. Accurate inference of tumor purity and absolute copy numbers from high-throughput sequencing data. Front. Genet. 2020;11:458. doi: 10.3389/fgene.2020.00458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yoshihara K., Shahmoradgoli M., Martínez E., Vegesna R., Kim H., Torres- Garcia W., Treviño V., Shen H., Laird P.W., Levine D.A., et al. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat. Commun. 2013;4:1–11. doi: 10.1038/ncomms3612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Racle J., de Jonge K., Baumgaertner P., Speiser D.E., Gfeller D. Simultaneous enumeration of cancer and immune cell types from bulk tumor gene expression data. eLife. 2017;6:e26476. doi: 10.7554/eLife.26476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Newman A.M., Steen C.B., Liu C.L., Gentles A.J., Chaudhuri A.A., Scherer F., Khodadoust M.S., Esfahani M.S., Luca B.A., Steiner D., et al. Determining cell type abundance and expression from bulk tissues with digital cytometry. Nat. Biotechnol. 2019;37:773–782. doi: 10.1038/s41587-019-0114-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li Y., Umbach D.M., Bingham A., Li Q.-J., Zhuang Y., Li L. Putative biomarkers for predicting tumor sample purity based on gene expression data. BMC Genomics. 2019;20:1–12. doi: 10.1186/s12864-019-6412-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Houseman E.A., Accomando W.P., Koestler D.C., Christensen B.C., Marsit C.J., Nelson H.H., Wiencke J.K., Kelsey K.T. DNA methylation arrays as surrogate measures of cell mixture distribution. BMC Bioinformatics. 2012;13:86. doi: 10.1186/1471-2105-13-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zheng X., Zhao Q., Wu H.-J., Li W., Wang H., Meyer C.A., Qin Q.A., Xu H., Zang C., Jiang P., et al. MethylPurify: tumor purity deconvolution and differential methylation detection from single tumor DNA methylomes. Genome Biol. 2014;15:1–13. doi: 10.1186/s13059-014-0419-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang N., Wu H.-J., Zhang W., Wang J., Wu H., Zheng X. Predicting tumor purity from methylation microarray data. Bioinformatics. 2015;31:3401–3405. doi: 10.1093/bioinformatics/btv370. [DOI] [PubMed] [Google Scholar]
- 39.Zheng X., Zhang N., Wu H.-J., Wu H. Estimating and accounting for tumor purity in the analysis of DNA methylation data from cancer studies. Genome Biol. 2017;18:1–14. doi: 10.1186/s13059-016-1143-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zack T.I., Schumacher S.E., Carter S.L., Cherniack A.D., Saksena G., Tabak B., Lawrence M.S., Zhang C.-Z., Wala J., Mermel C.H., et al. Pan-cancer patterns of somatic copy number alteration. Nat. Genet. 2013;45:1134–1140. doi: 10.1038/ng.2760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Akbani R., Ng P.K.S., Werner H.M., Shahmoradgoli M., Zhang F., Ju Z., Liu W., Yang J.-Y., Yoshihara K., Li J., et al. A pan-cancer proteomic perspective on the cancer genome atlas. Nat. Commun. 2014;5:1–15. doi: 10.1038/ncomms4887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cancer Genome Atlas Research Network Comprehensive molecular profiling of lung adenocarcinoma. Nature. 2014;511:543–550. doi: 10.1038/nature13385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen J., Yang H., Teo A.S.M., Amer L.B., Sherbaf F.G., Tan C.Q., Alvarez J.J.S., Lu B., Lim J.Q., Takano A., et al. Genomic landscape of lung adenocarcinoma in East Asians. Nat. Genet. 2020;52:177–186. doi: 10.1038/s41588-019-0569-6. [DOI] [PubMed] [Google Scholar]
- 44.Viray H., Coulter M., Li K., Lane K., Madan A., Mitchell K., Schalper K., Hoyt C., Rimm D.L. Automated objective determination of percentage of malignant nuclei for mutation testing. Appl. Immunohistochem. Mol. Morphol. 2014;22:363. doi: 10.1097/PAI.0b013e318299a1f6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hamilton P.W., Wang Y., Boyd C., James J.A., Loughrey M.B., Hougton J.P., Boyle D.P., Kelly P., Maxwell P., McCleary D., et al. Automated tumor analysis for molecular profiling in lung cancer. Oncotarget. 2015;6:27938. doi: 10.18632/oncotarget.4391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Azimi V., Chang Y.H., Thibault G., Smith J., Tsujikawa T., Kukull B., Jensen B., Corless C., Margolin A., Gray J.W. 2017 IEEE 14th International Symposium on Biomedical Imaging (ISBI 2017) IEEE; 2017. Breast cancer histopathology image analysis pipeline for tumor purity estimation; pp. 1137–1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pei Z., Cao S., Lu L., Chen W. Direct cellularity estimation on breast cancer histopathology images using transfer learning. Comput. Math. Methods Med. 2019;2019:3041250. doi: 10.1155/2019/3041250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rakhlin, A., Tiulpin, A., Shvets, A.A., Kalinin, A.A., Iglovikov, V.I., and Nikolenko, S.. (2019). Breast tumor cellularity assessment using deep neural networks. In Proceedings of the IEEE/CVF International Conference on Computer Vision Workshops, pages 0–0.
- 49.Greene C., O’Doherty E., Abdullahi Sidi F., Bingham V., Fisher N.C., Humphries M.P., Craig S.G., Harewood L., McQuaid S., Lewis C., et al. The potential of digital image analysis to determine tumor cell content in biobanked formalin-fixed, paraffin-embedded tissue samples. Biopreserv. Biobank. 2021;19:324–331. doi: 10.1089/bio.2020.0105. [DOI] [PubMed] [Google Scholar]
- 50.Campanella G., Hanna M.G., Geneslaw L., Miraflor A., Silva V.W.K., Busam K.J., Brogi E., Reuter V.E., Klimstra D.S., Fuchs T.J. Clinical-grade computational pathology using weakly supervised deep learning on whole slide images. Nat. Med. 2019;25:1301–1309. doi: 10.1038/s41591-019-0508-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tomita N., Abdollahi B., Wei J., Ren B., Suriawinata A., Hassanpour S. Attention-based deep neural networks for detection of cancerous and precancerous esophagus tissue on histopathological slides. JAMA Netw. open. 2019;2:e1914645. doi: 10.1001/jamanetworkopen.2019.14645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Oner, M.U., Lee, H.K., and Sung, W.-K.. (2020). Weakly supervised clustering by exploiting unique class count. In International Conference on Learning Representations.
- 53.Wilcoxon F. Breakthroughs in Statistics. Springer; 1992. Individual comparisons by ranking methods; pp. 196–202. [Google Scholar]
- 54.Oner M.U., Cheng Y.-C., Lee H.K., Sung W.-K. Training machine learning models on patient level data segregation is crucial in practical clinical applications. medRxiv. 2020 doi: 10.1101/2020.04.23.20076406. [DOI] [Google Scholar]
- 55.Meng X.-L., Rosenthal R., Rubin D.B. Comparing correlated correlation coefficients. Psychol. Bull. 1992;111:172. [Google Scholar]
- 56.Spencer D.H., Sehn J.K., Abel H.J., Watson M.A., Pfeifer J.D., Duncavage E.J. Comparison of clinical targeted next-generation sequence data from formalin-fixed and fresh-frozen tissue specimens. J. Mol. Diagn. 2013;15:623–633. doi: 10.1016/j.jmoldx.2013.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jamal-Hanjani M., Wilson G.A., McGranahan N., Birkbak N.J., Watkins T.B., Veeriah S., Shafi S., Johnson D.H., Mitter R., Rosenthal R., et al. Tracking the evolution of non–small-cell lung cancer. N. Engl. J. Med. 2017;376:2109–2121. doi: 10.1056/NEJMoa1616288. [DOI] [PubMed] [Google Scholar]
- 58.Gundem G., Van Loo P., Kremeyer B., Alexandrov L.B., Tubio J.M., Papaemmanuil E., Brewer D.S., Kallio H.M., Högnäs G., Annala M., et al. The evolutionary history of lethal metastatic prostate cancer. Nature. 2015;520:353. doi: 10.1038/nature14347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gerlinger M., Horswell S., Larkin J., Rowan A.J., Salm M.P., Varela I., Fisher R., McGranahan N., Matthews N., Santos C.R., et al. Genomic architecture and evolution of clear cell renal cell carcinomas defined by multiregion sequencing. Nat. Genet. 2014;46:225. doi: 10.1038/ng.2891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sottoriva A., Kang H., Ma Z., Graham T.A., Salomon M.P., Zhao J., Marjoram P., Siegmund K., Press M.F., Shibata D., et al. A big bang model of human colorectal tumor growth. Nat. Genet. 2015;47:209. doi: 10.1038/ng.3214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhai W., Lim T.K.-H., Zhang T., Phang S.-T., Tiang Z., Guan P., Ng M.-H., Lim J.Q., Yao F., Li Z., et al. The spatial organization of intra-tumour heterogeneity and evolutionary trajectories of metastases in hepatocellular carcinoma. Nat. Commun. 2017;8:4565. doi: 10.1038/ncomms14565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.McGranahan N., Swanton C. Biological and therapeutic impact of intratumor heterogeneity in cancer evolution. Cancer Cell. 2015;27:15–26. doi: 10.1016/j.ccell.2014.12.001. [DOI] [PubMed] [Google Scholar]
- 63.Efron B. Breakthroughs in Statistics. Springer; 1992. Bootstrap methods: another look at the jackknife; pp. 569–593. [Google Scholar]
- 64.Yu K.-H., Zhang C., Berry G.J., Altman R.B., Ré C., Rubin D.L., Snyder M. Predicting non-small cell lung cancer prognosis by fully automated microscopic pathology image features. Nat. Commun. 2016;7:1–10. doi: 10.1038/ncomms12474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sozzi G., Conte D., Leon M., Cirincione R., Roz L., Ratcliffe C., Roz E., Cirenei N., Bellomi M., Pelosi G., et al. Quantification of free circulating DNA as a diagnostic marker in lung cancer. J. Clin. Oncol. 2003;21:3902–3908. doi: 10.1200/JCO.2003.02.006. [DOI] [PubMed] [Google Scholar]
- 66.Coudray N., Ocampo P.S., Sakellaropoulos T., Narula N., Snuderl M., Fenyö D., Moreira A.L., Razavian N., Tsirigos A. Classification and mutation prediction from non–small cell lung cancer histopathology images using deep learning. Nat. Med. 2018;24:1559–1567. doi: 10.1038/s41591-018-0177-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Fu Y., Jung A.W., Torne R.V., Gonzalez S., Vöhringer H., Shmatko A., Yates L.R., Jimenez-Linan M., Moore L., Gerstung M. Pan-cancer computational histopathology reveals mutations, tumor composition and prognosis. Nat. Cancer. 2020;1:1–11. doi: 10.1038/s43018-020-0085-8. [DOI] [PubMed] [Google Scholar]
- 68.Hanahan D., Coussens L.M. Accessories to the crime: functions of cells recruited to the tumor microenvironment. Cancer Cell. 2012;21:309–322. doi: 10.1016/j.ccr.2012.02.022. [DOI] [PubMed] [Google Scholar]
- 69.Junttila M.R., de Sauvage F.J. Influence of tumour micro-environment heterogeneity on therapeutic response. Nature. 2013;501:346. doi: 10.1038/nature12626. [DOI] [PubMed] [Google Scholar]
- 70.Chen K.H., Boettiger A.N., Moffitt J.R., Wang S., Zhuang X. Spatially resolved, highly multiplexed RNA profiling in single cells. Science. 2015;348 doi: 10.1126/science.aaa6090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Svensson V., Teichmann S.A., Stegle O. SpatialDE: identification of spatially variable genes. Nat. Methods. 2018;15:343–346. doi: 10.1038/nmeth.4636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Moffitt J.R., Bambah-Mukku D., Eichhorn S.W., Vaughn E., Shekhar K., Perez J.D., Rubinstein N.D., Hao J., Regev A., Dulac C., et al. Molecular, spatial, and functional single-cell profiling of the hypothalamic preoptic region. Science. 2018;362:eaau5324. doi: 10.1126/science.aau5324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Stuart T., Butler A., Hoffman P., Hafemeister C., Papalexi E., Mauck W.M., III, Hao Y., Stoeckius M., Smibert P., Satija R. Comprehensive integration of single-cell data. Cell. 2019;177:1888–1902. doi: 10.1016/j.cell.2019.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Deng J., Dong W., Socher R., Li L.-J., Li K., Fei-Fei L.. 2009 IEEE conference on computer vision and pattern recognition. Ieee; 2009. ImageNet: A Large-Scale Hierarchical Image Database; pp. 248–255. [Google Scholar]
- 75.Caiado F., Silva-Santos B., Norell H. Intra-tumour heterogeneity–going beyond genetics. FEBS J. 2016;283:2245–2258. doi: 10.1111/febs.13705. [DOI] [PubMed] [Google Scholar]
- 76.Lazar A.J., McLellan M.D., Bailey M.H., Miller C.A., Appelbaum E.L., Cordes M.G., Fronick C.C., Fulton L.A., Fulton R.S., Mardis E.R., et al. Comprehensive and integrated genomic characterization of adult soft tissue sarcomas. Cell. 2017;171:950–965. doi: 10.1016/j.cell.2017.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.L020 standard operating procedure (SOP) for sectioning and portioning frozen tissue samples in logistics. 2020. https://brd.nci.nih.gov/brd/sop/show/1447
- 78.H006 standard operating procedure (SOP) for sectioning frozen tissue samples in histology. 2020. https://brd.nci.nih.gov/brd/sop/show/1442
- 79.H001 standard operating procedure (SOP) for hematoxylin and eosin (H&E) staining coverslipping. 2020. https://brd.nci.nih.gov/brd/sop/show/1421
- 80.L016 standard operating procedure (SOP) for preparing frozen tissue for molecular analysis. 2020. https://brd.nci.nih.gov/brd/sop/show/1445
- 81.Oner M.U., Kye-Jet J.M.S., Lee H.K., Sung W.-K. Studying the effect of MIL pooling filters on MIL tasks. arXiv. 2020 https://arxiv.org/abs/2006.01561 arXiv:2006.01561. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
All TCGA datasets are publicly available. Manifest files are provided with original code to download H&E-stained digital histopathology slides using GDC Data Transfer Tool. Genomic tumor purity values were downloaded from https://gdc.cancer.gov/about-data/publications/pancanatlas under filename TCGA_mastercalls.abs_ tables_JSedit.fixed.txt. They are also given in Data S1. For the Singapore cohort, genomic tumor purity values and representative histological images are publicly available from OncoSG (https://src.gisapps.org/OncoSG/) under dataset Lung Adenocarcinoma (GIS, 2019).
-
•
All original code has been deposited at Zenodo under the https://doi.org/10.5281/zenodo.5606981 and is publicly available as of the date of publication. The repository provides a detailed step-by-step explanation, from downloading H&E-stained digital histopathology slides to obtaining spatially resolved tumor purity maps (SRTPMs).
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.