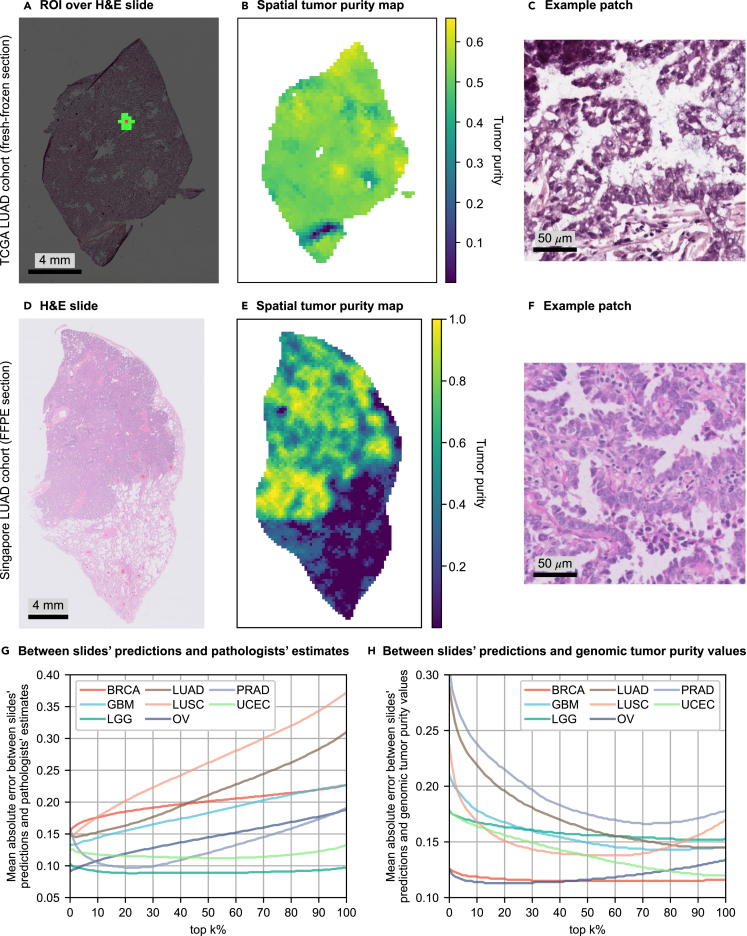
Figure 4.
Incorrect size and selection of ROI might cause overestimation in percentage tumor nuclei estimates
(A–H) For a slide of a fresh-frozen section in the TCGA LUAD cohort, (A) shows the ROI centered on a patch and consisting of 16 closest patches to that particular patch (≈1 mm2 at the specimen level). Tumor purity corresponding to the patch is predicted over the ROI. (B) The tumor purity map for all patches within the slide. Similarly, (D) shows a slide of a FFPE section in the LUAD_SG cohort, and (E) shows its corresponding tumor purity map. (C) and (F) show example patches cropped from cancerous regions in the slides shown in (A) and (D), respectively. (G and H) To investigate the effect of the size and selection of ROI on pathologists' percentage tumor nuclei estimates, we conducted error analyses over the slides' tumor purity values by gradually extending the ROI. We calculated the slide's tumor purity as the average of top-k% of the patches with the highest scores (k = 0, ···, 100) in the tumor purity map (k = 0: the patch with the highest tumor purity). In different cohorts, we plotted mean absolute error versus top-k% of the patches for error analyses between slides' predictions and pathologists' percentage tumor nuclei estimates in (G) and slides' predictions and genomic tumor purity values in (H). See also Figures S4 and S6–S10.