Figure 6:
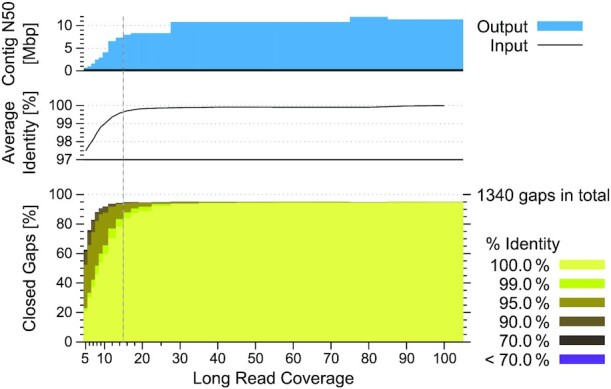
Coverage analysis by applying DENTIST to Drosophila with varying coverage of simulated PacBio CLR reads. The figure compares contig NG50 (top), average identity of the closed gaps (middle), and the number of closed gaps together with a breakdown of their sequence identity (bottom). The number of closed gaps starts to plateau above a read coverage of ∼15× (indicated by a vertical dashed line), which is significantly less than the recommended coverage of ∼60× required for de novo genome assembly.
