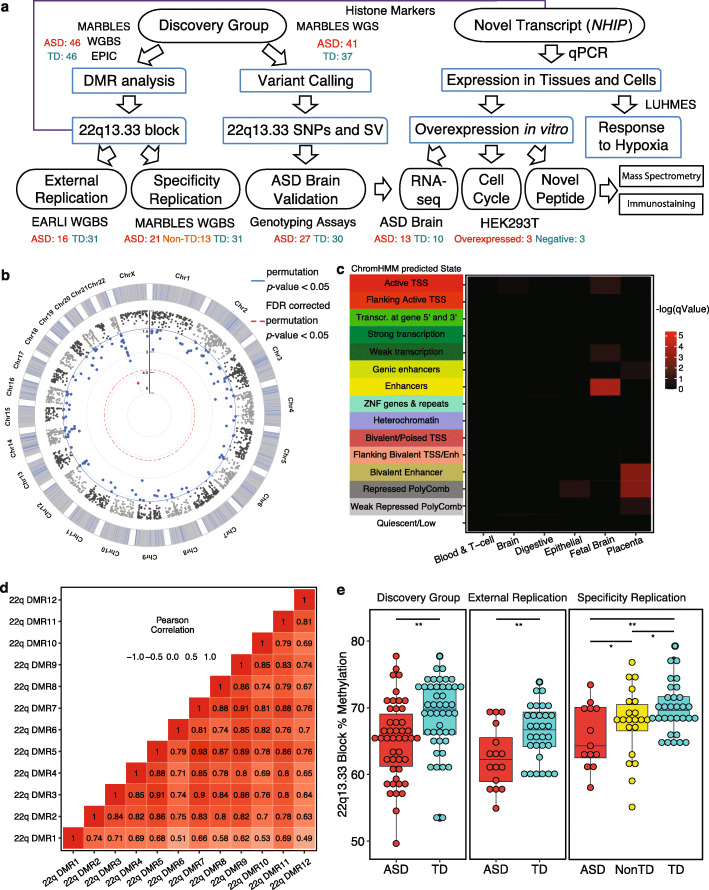
Fig. 1.
ASD-associated DMRs are enriched at fetal brains enhancers and a co-methylated block at 22q13.33 replicates across studies and platforms. A Schematic of the experimental design for discovery of ASD DMRs, replication of the co-methylated 22q13.33 locus, genetic associations, and functional follow-up of a novel gene (NHIP). B Circular Manhattan plot of the epigenome-wide association of DNA methylation in placenta with ASD diagnosis at 36 months. Results are represented as DMR association test results (− log10(p)). Significant thresholds are blue for permutation p value < 0.05, red for FDR-adjusted permutation p value < 0.05, and gray for nonsignificant. C 134 ASD DMRs (permutation p value < 0.05) tested for enrichment within chromatin states defined by Epigenome Roadmap ChromHMM [35]. Each row represents a different ChromHMM state and each column a single tissue type, with the heatmap plotting the − log10(q-value) significance of ASD DMR enrichment. D Correlation matrix of methylation levels using the Pearson correlation coefficient for the 12 DMRs located in the 22q13.33 hypomethylated block. E Smoothed methylation values were averaged over the 22q13.33 hypomethylated block (y-axis) and compared across diagnosis groups (x-axis). In the discovery group, ASD samples had significantly lower methylation than TD samples (MARBLES, HiSeq X, ASD n = 46, TD n = 46) (p value = 0.002). The same result and direction were observed in the external replication group (EARLI, HiSeq 2500, ASD n = 16, TD n = 31) (p value = 0.009). For the specificity replication group (MARBLES, NovaSeq, ASD n = 21, Non-TD n = 13, TD n = 31), ASD methylation levels were also significantly lower than both TD (p value = 0.005) and Non-TD (p value = 0.049), while Non-TD was significantly lower than TD samples (p value = 0.050) by Mann-Whitney-Wilcoxon. Box plot center lines, box limits, and whiskers represented median, interquartile range, and minimum and maximum values, respectively