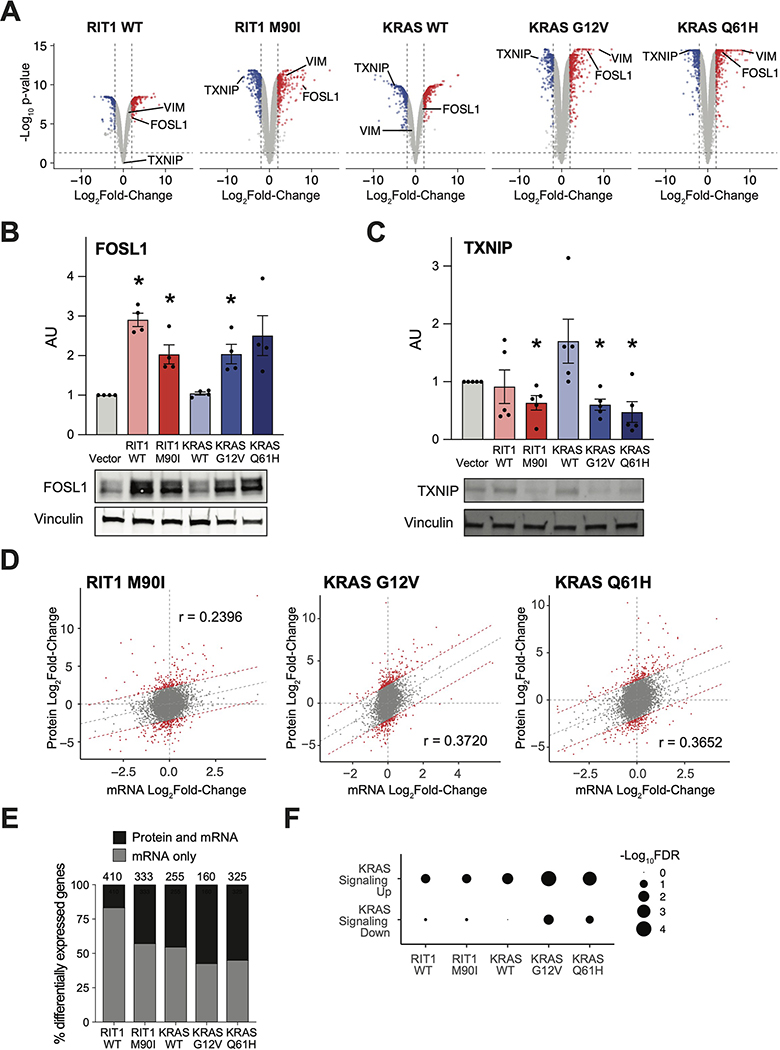
Figure 2. Quantitative proteomic and transcriptomic profiling identifies similarity of oncogenic RIT1 and KRAS signaling.
(A) Volcano plots of global proteome data from isogenic AALE cells showing the Log2Fold-Change in protein abundance in each cell line compared to vector control cells. The y-axis displays the negative Log10(P-value) calculated from a one sample moderated t-test with multiple hypothesis correction by the Benjamini-Hochberg method. Data shown are generated from 2 to 4 technical LC-MS/MS replicates per cell line generated from N=2 biological replicates per cell line. (B and C) Western blotting of FOSL1 (B) and TXNIP (C) protein abundance in RIT1-variant and KRAS-variant AALE cells. Quantification of N = 4 (B) and 5 (C) independent experiments alongside a representative blot for each. * P < 0.05, compared to vector condition, by two-tailed t-test. (D) Global proteome-transcriptome correlation analysis for RIT1-variant and KRAS-variant AALE cell lines. Gray dashed line indicates linear regression between Log2Fold-Change of mRNA to respective Log2Fold-Change of protein. Red denotes genes outside the 95% prediction interval. r = Pearson correlation coefficient. Data shown are generated from 2 to 4 technical LC-MS/MS replicates per cell line generated from N=2 biological replicates per cell line. N = 3 RNA-seq replicates for each isogenic cell line. (E) In RIT1- or KRAS-expressing AALE cell lines compared to vector control AALE cells, the proportion of genes that are differentially expressed at the mRNA level (log2Fold-Change > 1, FDR < 0.05) which are also differentially abundant at the protein level (log2Fold-Change > 1, adjusted P-value < 0.05). Number of total differentially expressed mRNA transcripts is shown above the bar for each cell line. N is as described in (D). (F) Gene set enrichment analysis of differentially expressed gene transcripts between KRAS or RIT1 AALE cell lines and vector controls using GOseq (56). mSigDB hallmark gene sets specific to KRAS signaling are shown. Circle size = −Log10FDR of enrichment significance determined by GOseq. Analysis performed with N = 3 RNA-seq replicates per isogenic cell line.