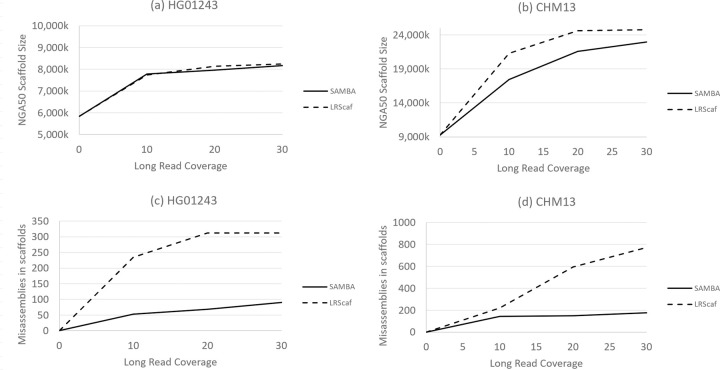
Fig 2. Contiguity and accuracy results for SAMBA and LRScaf on scaffolding of human assemblies CHM13 (CHM13-WashU) and HG01243 (HG01243-IP) using varied coverage of Oxford Nanopore Technologies (ONT) ultralong reads.
The x-axis shows coverage in the ONT reads. All assemblies were evaluated by Quast using the CHM13 v1.1 assembly as the reference for CHM13, and the PR1 assembly (Zimin et al., 2021) for HG01243. Panels (a) and (b) compare NG50 sizes for the two different human assemblies, while (c) and (d) compare the number of mis-assemblies. Both scaffolders produced similar NG50 sizes for HG01243, while LRScaf produced bigger scaffolds for CHM13 data. SAMBA had fewer mis-assemblies in both genomes at all depths of coverage.