Figure 4.
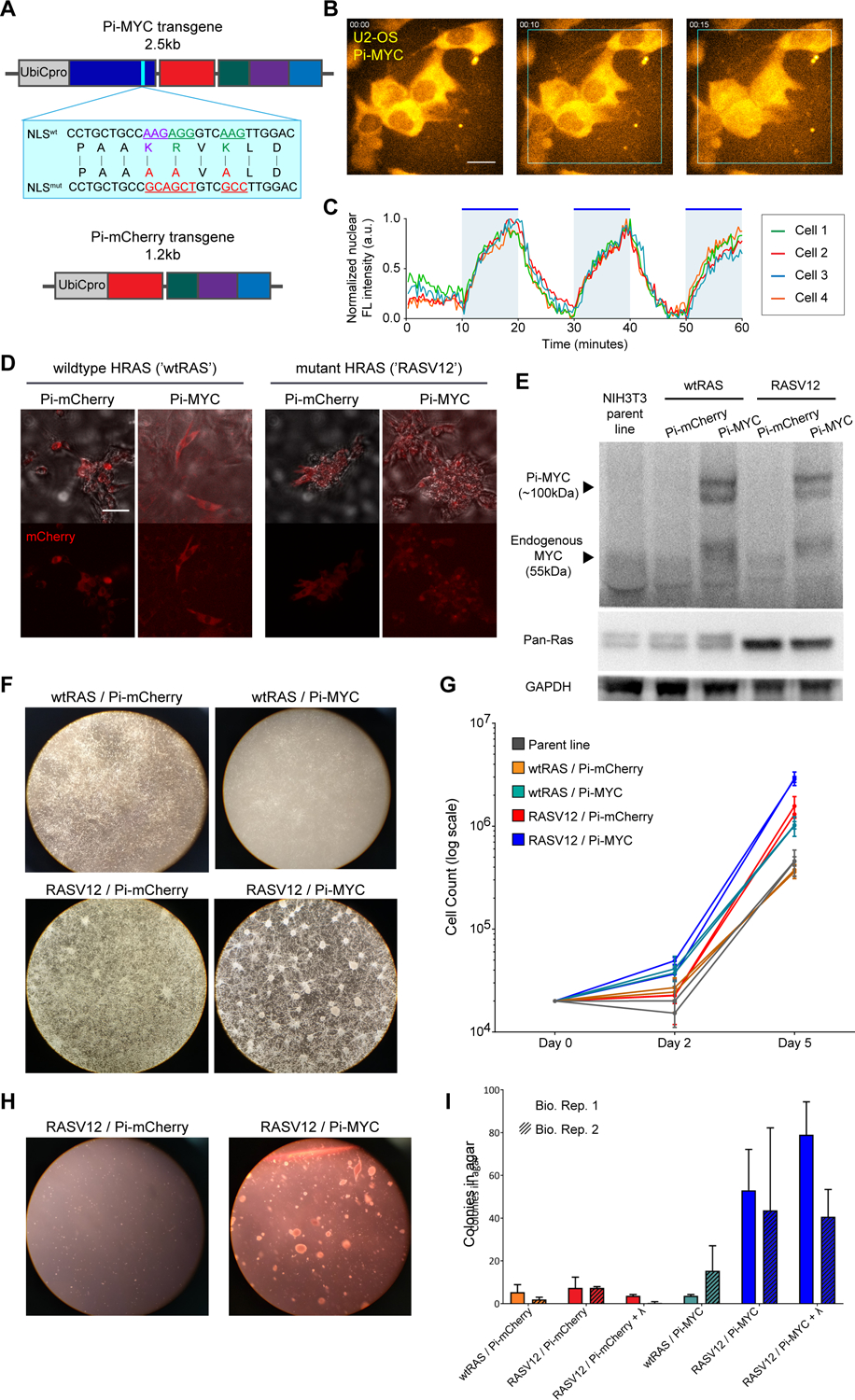
Photo-inducible MYC displays oncogenic capability in NIH3T3 cells.
(A) Schematic of Pi-MYC. The transgene consists of the human Ubiquitin C promoter (UbiCpro), c-myc exons 2 and 3 (MYC), alanine mutations in the native NLS (blue inset shows changed residues), mCherry, a nuclear export signal (NES), and the LOV2 domain followed by the wildtype c-myc NLS sequence. The Pi-mCherry control transgene contains all domains except MYC.
(B) Pi-MYC stable expression in U2OS cells. Scale bar = 15μm. In the absence of irradiating light, the NES allows Pi-MYC retention in the cytoplasm. Upon irradiation with blue-green wavelengths of light (450–500nm), the LOV2 domain exposes the enclosed NLS and allows Pi-MYC to be imported into the nucleus (indicated by white triangles). Pi-mCherry operates via the same mechanism.
(C) Quantification of nuclear fluorescence intensity from four cells numbered in (B) over a 1-hour time series. The field of view was subjected to alternating 10-minute periods of activating light indicated by the blue regions in the plot.
(D) DIC merge and fluorescence images of NIH3T3 fibroblasts stably expressing Pi-mCherry or Pi-MYC (visualized with mCherry) in a background of wildtype HRAS or V12 mutant. Scale bar = 50μm.
(E) Western blot of MYC and RAS expression in the NIH3T3 stable lines.
(F) Growth and focus formation in monolayer culture of four NIH3T3 lines.
(G) Quantification of growth rates, two biological replicates per cell line. Error bars are SD of three technical replicates.
(H) Colony formation in soft agar of Pi-mCherry and Pi-MYC stable lines in the RAS mutant background.
(I) Quantification of colonies visible after 2 weeks growth in soft agar of the four stable NIH3T3 lights, and the RASV12 lines cultured with 24 hours light before embedding in agar. Error bars are SD of two biological replicates.
