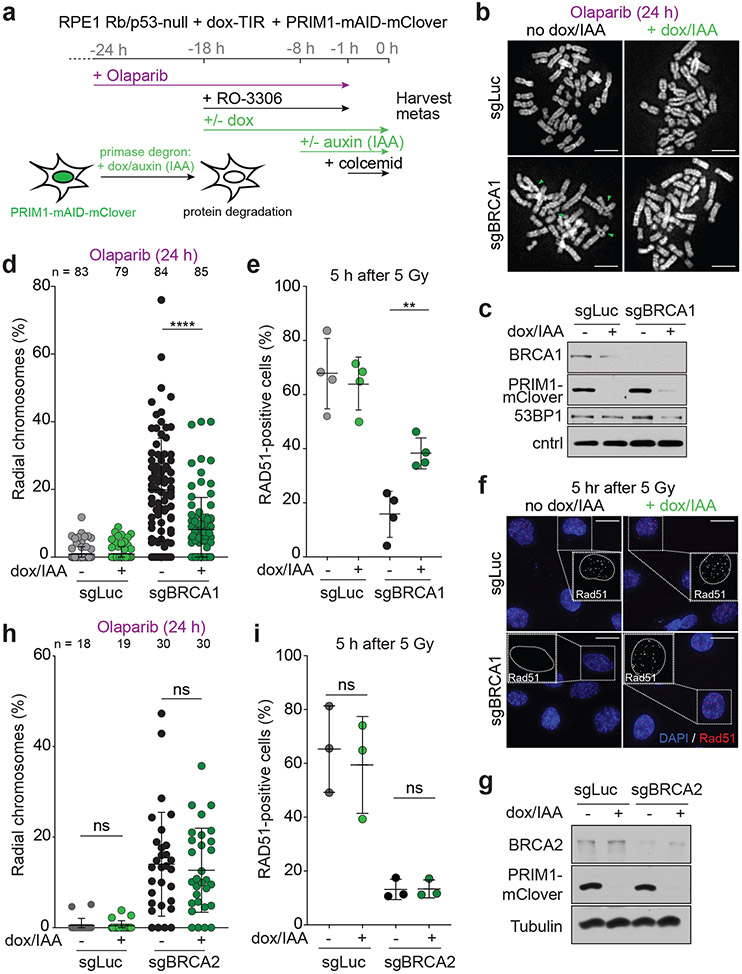
Fig. 1. Primase promotes radials and blocks RAD51 loading in BRCA1-deficient cells.
a, Schematic of the timeline of auxin-induced degradation of PRIM1 in G2-arrested RPE1 cells treated with PARPi. b, Representative images of DAPI-stained metaphase spreads from RPE1 PRIM1-mAID-mClover cells with the indicated treatments. Green arrows: aberrant radial chromosomes. Scale bars: 5 μm. c, Immunoblot for BRCA1 and GFP (mClover-PRIM1) in cells as in a treated with control (sgLuc) or BRCA1 (sgBRCA1) bulk CRISPR KO. ctrl, non-specific band from GFP blot. d, Quantification of the percent of chromosomes involved in radial structures. Number of metaphases analyzed per condition (n) is indicated. e, Quantification of the percent of RAD51-positive cells (with 10 or more RAD51 foci per nucleus) 5 h after 5 Gy. n = four independent experiments. f, Representative images of IF for RAD51 in cells as in e with the indicated treatments. Scale bars: 20 μm. g, Immunoblot for BRCA2 and GFP (mClover-PRIM1) in cells as in a treated with control (sgLuc) or BRCA2 (sgBRCA2) bulk CRISPR KO. h, Quantification of the percent of chromosomes involved in radial structures (as in d). Number of metaphases analyzed per condition (n) is indicated. i, Quantification of the percent of RAD51-positive cells as in e, in the indicated cells. n = three independent experiments. Data shown in b-d, and f-i are representative of three independent experiments. All statistical analysis based on two-tailed Welch’s t-test. *, p<0.05; **, p<0.01; ***, p<0.001; ****, p<0.0001; ns, not significant. All means are indicated with center bars and SDs with error bars.