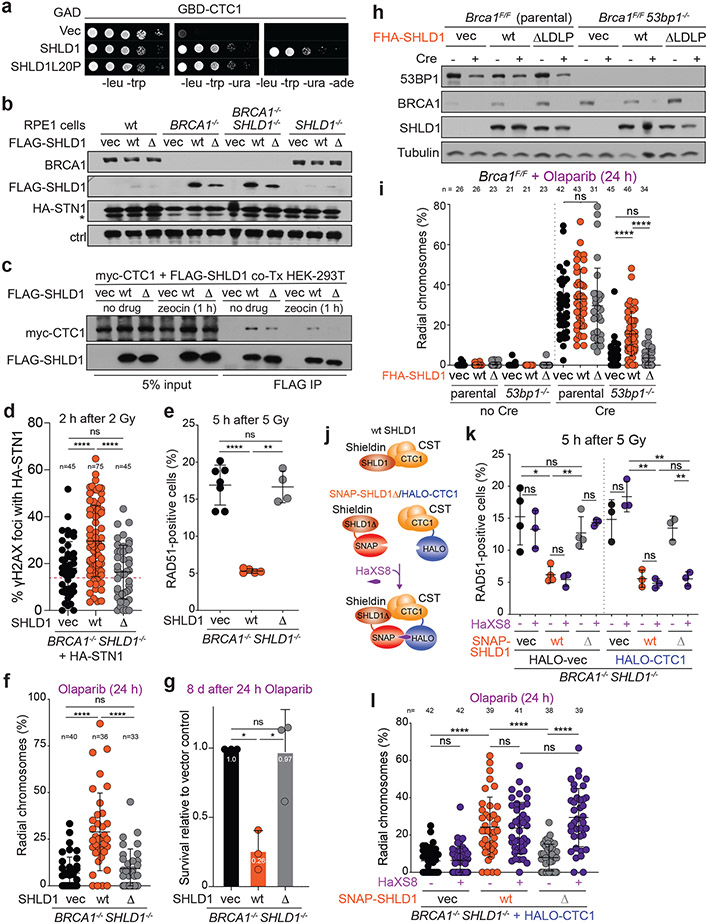
Fig. 5. Shieldin function in BRCA1-deficient cells depends on the SHLD1-CTC1 interaction.
a, Yeast-two hybrid assay demonstrating lack of interaction between human SHLD1L20P and CTC1 proteins. Colony growth on permissive (-leucine, -tryptophan, -uracil), but not selective (-leucine, -tryptophan, -uracil, -adenine) media indicates lack of interaction. b, Immunoblots in the indicated cells. SHLD1Δ (Δ) has a deletion of aa 18-21. ctrl, non-specific band from STN1 blot. a and b are representative of two independent experiments. c, Immunoprecipitation of FLAG-SHLD1 and immunoblot for myc-CTC1 co-expressed in 293T cells. Representative of four independent experiments. d, Quantification of IR-induced γH2AX foci with HA-STN1 signal in BRCA1/SHLD1 DKO cells as in b. Number of nuclei (n, each represented by a dot) pooled from three independent experiments is indicated. Red dotted line: the average background level due to randomly overlapping γH2AX and HA foci (see Materials and Methods). e, Quantification of the percent of RAD51-positive cells in irradiated BRCA1/SHLD1 DKO RPE1 cells complemented with the indicated FLAG-SHLD1 construct or an empty vector (vec). Each dot represents an independent experiment (n = four-seven experiments involving >60 cells each). f, Quantification of the percent of chromosomes in radial structures in cells as in e. Number of metaphase spreads (n, each represented by a dot) pooled from three independent experiments is indicated. g, Quantification of colony formation by BRCA1/SHLD1 DKO cells as in e treated with 5 μM Olaparib for 24 h. Survival after PARPi was compared to undrugged cells and normalized to empty vector. n = three independent experiments. h, Immunoblot for BRCA1, 53BP1, and SHLD1 detecting FHA-tagged SHLD1 in the indicated cells. Representative of three independent experiments. i, Quantification of the percent of chromosomes in radial structures in the indicated MEFs expressing FHA-SHLD1Δ (Δ). Empty vector (vec) and FHA-SHLD1 (wt) conditions from Fig. 4b are provided again here. Number of metaphase spreads (n, each represented by a dot) pooled from three independent experiments is indicated. j, Schematic of HaXS8-induced dimerization of SNAP-SHLD1Δ with HALO-CTC1. k, Quantification of RAD51-positive cells in irradiated BRCA1/SHLD1 DKO cells, complemented with the indicated SNAP-SHLD1 or empty vector, and HALO-CTC1 or empty vector, then treated with HaXS8 or vehicle prior to irradiation. n = three or four independent experiments as indicated. l, Quantification of the percent of chromosomes in radial structures in BRCA1/SHLD1 DKO cells with the indicated treatments. Number of metaphase spreads (n, each represented by a dot) pooled from three independent experiments is indicated. Statistical analyses as in Fig. 1. All means are indicated with center bars and SDs with error bars.